Abstract
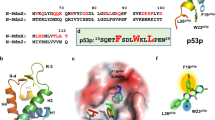
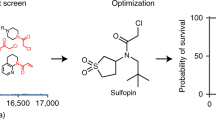
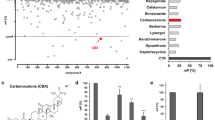
Here, we report the fragment-based discovery of BI-9321, a potent, selective and cellular active antagonist of the NSD3-PWWP1 domain. The human NSD3 protein is encoded by the WHSC1L1 gene located in the 8p11-p12 amplicon, frequently amplified in breast and squamous lung cancer. Recently, it was demonstrated that the PWWP1 domain of NSD3 is required for the viability of acute myeloid leukemia cells. To further elucidate the relevance of NSD3 in cancer biology, we developed a chemical probe, BI-9321, targeting the methyl-lysine binding site of the PWWP1 domain with sub-micromolar in vitro activity and cellular target engagement at 1 µM. As a single agent, BI-9321 downregulates Myc messenger RNA expression and reduces proliferation in MOLM-13 cells. This first-in-class chemical probe BI-9321, together with the negative control BI-9466, will greatly facilitate the elucidation of the underexplored biological function of PWWP domains.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The authors declare that the data supporting the findings of this study are available within the publication and its Supplementary Information files or have been deposited in the RCSB Protein Data Bank (PDB, http://www.rcsb.org) with the following accession numbers: selenomethionine labeled (6G3P); unlabeled (6G3T); X-ray structure of NSD3-PWWP1 in complex with compound 3 (6G24); X-ray structure of NSD3-PWWP1 in complex with compound 4 (6G25); X-ray structure of NSD3-PWWP1 in complex with compound 5 (6G27); X-ray structure of NSD3-PWWP1 in complex with compound 6 (6G29); X-ray structure of NSD3-PWWP1 in complex with compound 8 (6G2B); X-ray structure of NSD3-PWWP1 in complex with compound 9 (6G2C); X-ray structure of NSD3-PWWP1 in complex with compound 13 (6G2E); X-ray structure of NSD3-PWWP1 in complex with compound 16 (6G2F); X-ray structure of NSD3-PWWP1 in complex with compound BI-9321 (6G2O).
References
Allis, C. D. & Jenuwein, T. The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 17, 487–500 (2016).
Bannister, A. J. & Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 21, 381–395 (2011).
Arrowsmith, C. H., Bountra, C., Fish, P. V., Lee, K. & Schapira, M. Epigenetic protein families: a new frontier for drug discovery. Nat. Rev. Drug Discov. 11, 384–400 (2012).
Brown, P. J. & Müller, S. Open access chemical probes for epigenetic targets. Future Med. Chem. 7, 1901–1917 (2015).
Huston, A., Arrowsmith, C. H., Knapp, S. & Schapira, M. Probing the epigenome. Nat. Chem. Biol. 11, 542 (2015).
Yang, L., Rau, R. & Goodell, M. A. DNMT3A in haematological malignancies. Nat. Rev. Cancer 15, 152 (2015).
Vougiouklakis, T., Hamamoto, R., Nakamura, Y. & Saloura, V. The NSD family of protein methyltransferases in human cancer. Epigenomics 7, 863–874 (2015).
Kang, D. et al. The histone methyltransferase Wolf–Hirschhorn syndrome candidate 1-like 1 (WHSC1L1) is involved in human carcinogenesis. Genes, Chromosome. Cancer 52, 126–139 (2013).
Angrand, P. O. et al. NSD3, a new SET domain-containing gene, maps to 8p12 and is amplified in human breast cancer cell lines. Genomics 74, 79–88 (2001).
Shen, C. et al. NSD3-short is an adaptor protein that couples BRD4 to the CHD8 chromatin remodeler. Mol. Cell 60, 847–859 (2015).
Gelsi-Boyer, V. et al. Comprehensive profiling of 8p11-12 amplification in breast cancer. Mol. Cancer Res. 3, 655 (2005).
Wu, H. et al. Structural and histone binding ability characterizations of human PWWP domains. PLoS ONE 6, e18919 (2011).
Qin, S. & Min, J. Structure and function of the nucleosome-binding PWWP domain. Trends Biochem. Sci. 39, 536–547 (2014).
Vezzoli, A. et al. Molecular basis of histone H3K36me3 recognition by the PWWP domain of Brpf1. Nat. Struct. Mol. Biol. 17, 617–619 (2010).
Hubbard, R. E. Fragment approaches in structure-based drug discovery. J. Synchrotron Rad. 15, 227–230 (2008).
Baurin, N. et al. Design and characterization of libraries of molecular fragments for use in NMR screening against protein targets. J. Chem. Inf. Comp. Sci. 44, 2157–2166 (2004).
Bergner, A. & Parel, S. P. Hit expansion approaches using multiple similarity methods and virtualized query structures. J. Chem. Inf. Modeling 53, 1057–1066 (2013).
Hosmane, R. S. & Liebman, J. F. Paradoxes and paradigms: why is quinoline less basic than pyridine or isoquinoline? A classical organic chemical perspective. Struct. Chem. 20, 693–697 (2009).
Fang, R. et al. Human LSD2/KDM1b/AOF1 regulates gene transcription by modulating intragenic H3K4me2 methylation. Mol. Cell 39, 222–233 (2010).
Machleidt, T. et al. NanoBRET—a novel BRET platform for the analysis of protein–protein interactions. ACS Chem. Bio. 10, 1797–1804 (2015).
Jafari, R. et al. The cellular thermal shift assay for evaluating drug target interactions in cells. Nat. Protoc. 9, 2100–2122 (2014).
Dart, M. L. et al. Homogeneous assay for target engagement utilizing bioluminescent thermal shift. ACS Med. Chem. Lett. 9, 546–551 (2018).
Philpott, M. et al. Assessing cellular efficacy of bromodomain inhibitors using fluorescence recovery after photobleaching. Epigenetics & Chromatin 7, 14 (2014).
Frye, S. V. The art of the chemical probe. Nat. Chem. Biol. 6, 159–161 (2010).
Ross, A. & Senn, H. Automation of measurements and data evaluation in biomolecular NMR screening. Drug Discov. 6, 583–593 (2001).
Mayer, M. & Meyer, B. Characterization of ligand binding by saturation transfer difference NMR Spectroscopy. Angew. Chem. Int. Edn 38, 1784–1788 (1999).
Nietlispach, D. Suppression of anti-TROSY lines in a sensitivity enhanced gradient selection TROSY scheme. J. Biomol. NMR 31, 161–166 (2005).
Pervushin, K., Riek, R., Wider, G. & Wüthrich, K. Attenuated T(2) relaxation by mutual cancellation of dipole–dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc. Natl Acad. Sci. USA 94, 12366–12371 (1997).
Grzesiek, S., Stahl, S. J., Wingfield, P. T. & Bax, A. The CD4 determinant for downregulation by HIV-1 Nef Directly Binds to Nef. Mapping of the Nef Binding Surface by NMR. Biochem. 35, 10256–10261 (1996).
Peng, C., Unger, S. W., Filipp, F. V., Sattler, M. & Szalma, S. Automated evaluation of chemical shift perturbation spectra: new approaches to quantitative analysis of receptor-ligand interaction NMR spectra. J. Biomol. NMR 29, 491–504 (2004).
Martin, L. J. et al. Structure-sased design of an in vivo active selective BRD9 inhibitor. J. Med. Chem. 59, 4462–4475 (2016).
Willett, P. Combination of similarity rankings using data fusion. J. Chem. Inf. Mod. 53, 1–10 (2013).
Hert, J. et al. New methods for ligand-based virtual screening: use of data fusion and machine learning to enhance the effectiveness of similarity searching. J. Chem. Inf. Mod. 46, 462–470 (2006).
Hawkins, P. C. D., Skillman, A. G. & Nicholls, A. Comparison of shape-matching and docking as virtual screening tools. J. Med. Chem. 50, 74–82 (2007).
Cheeseright, T. J., Mackey, M. D., Melville, J. L. & Vinter, J. G. FieldScreen: virtual screening using molecular fields. Application to the DUD data set. J. Chem. Inf. Mod. 48, 2108–2117 (2008).
Cox, J. et al. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Prot. 13, 2513–2526 (2014).
Yu, W. et al. A scintillation proximity assay for histone demethylases. Anal. Biochem. 463, 54–60 (2014).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010).
Vonrhein, C. et al. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr. D 67, 293–302 (2011).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of COOT. Acta Crystallogr. D 66, 486–501 (2010).
Collaborative. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994).
Robers, M. B. et al. Target engagement and drug residence time can be observed in living cells with BRET. Nat. Comm. 6, 10091–10091 (2015).
Acknowledgements
The Structural Genomics Consortium is a registered charity (no. 1097737) that receives funds from AbbVie; Bayer Pharma AG; Boehringer Ingelheim; Canada Foundation for Innovation; Eshelman Institute for Innovation; Genome Canada; Innovative Medicines Initiative (EU/EFPIA) (ULTRA-DD grant no. 115766); Janssen; Merck & Co.; Novartis Pharma AG; Ontario Ministry of Economic Development and Innovation; Pfizer; São Paulo Research Foundation-FAPESP; Takeda and the Wellcome Trust. We thank the Expose team for data collection at the Swiss Light Source beamlines X06SA and X06DA. We thank D. Daniels, M. Robers and C. Corona from Promega for advising on the NanoBRET and target engagement assays, and acknowledge the Natural Sciences and Engineering Research Council of Canada (NSERC) for a postdoctoral fellowship awarded to D.D.
Author information
Authors and Affiliations
Contributions
J.B. and U.R. supervised the chemistry team. U.R., T.W., S.Z. and D.B. designed synthetic strategies. D.D., R.A.N., M.S., M. Petronczki, D.B.-L., M.V., K.V.M.H., C.R.V. and M. Pearson designed biological experiments and supervised the biology teams. M.Z. supervised the DSF, NMR measurements of the FBS screening. A.Z. supervised protein production and ITC experiments. J.B., B.M. and A.W.-P. performed structural analysis. K.R., S.K. and S.W. performed SPR assays. O.F., F.L. and A.A.-H. performed selectivity assays. A.A.-H. contributed to performing SPR and ITC experiments. C.M.R. performed FRAP assays. M.M.S. performed NanoBRET assays. D.D. and M.C. performed cellular experiments. C.W. synthesized tracer ligands. M.M. and H.B. performed analytics and wrote the Supplementary Note. N.M. performed quantitative proteomics experiments. H.A. performed quantitative tandem mass spectroscopy. C.R., A.H. and T.K. performed biological experiments. B.S., D.H. and T.G. performed biochemical assays. X.-L.C. and J.E.F. provided Compchem support. G.B. supervised the collaboration with SGC. P.J.B. managed the project for SGC. C.H.A. provided supervision and funding. D.B.M. was responsible for the medicinal chemistry strategy. J.B., D.D., R.A.N. and U.R. prepared the manuscript with input from all authors.
Corresponding author
Ethics declarations
Competing interests
J.B., U.R., R.A.N., M. Petronczki, M.Z., N.M., K.R., A.Z., M.M., T.W., S.Z., H.A., H.B., C.M.R., A.H., T.K., M.C., B.S., S.W., D.H., X.-L.C., J.E.F., B.M., A.W.-P., T.G., G.B., M. Pearson and D.B.M. are employees of Boehringer Ingelheim.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Tables 1–13, Supplementary Figs. 1–13
Supplementary Note
Synthetic procedures
Rights and permissions
About this article
Cite this article
Böttcher, J., Dilworth, D., Reiser, U. et al. Fragment-based discovery of a chemical probe for the PWWP1 domain of NSD3. Nat Chem Biol 15, 822–829 (2019). https://doi.org/10.1038/s41589-019-0310-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-019-0310-x
This article is cited by
-
Evaluating the use of absolute binding free energy in the fragment optimisation process
Communications Chemistry (2022)
-
The role of NSD1, NSD2, and NSD3 histone methyltransferases in solid tumors
Cellular and Molecular Life Sciences (2022)
-
A chemical probe targeting the PWWP domain alters NSD2 nucleolar localization
Nature Chemical Biology (2022)
-
Epigenetics and beyond: targeting writers of protein lysine methylation to treat disease
Nature Reviews Drug Discovery (2021)
-
Targeting non-bromodomain chromatin readers
Nature Structural & Molecular Biology (2019)