Abstract
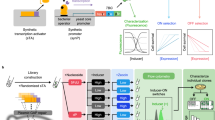
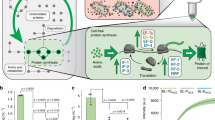
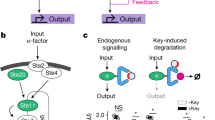
To maximize a desired product, metabolic engineers typically express enzymes to high, constant levels. Yet, permanent pathway activation can have undesirable consequences including competition with essential pathways and accumulation of toxic intermediates. Faced with similar challenges, natural metabolic systems compartmentalize enzymes into organelles or post-translationally induce activity under certain conditions. Here we report that optogenetic control can be used to extend compartmentalization and dynamic control to engineered metabolisms in yeast. We describe a suite of optogenetic tools to trigger assembly and disassembly of metabolically active enzyme clusters. Using the deoxyviolacein biosynthesis pathway as a model system, we find that light-switchable clustering can enhance product formation six-fold and product specificity 18-fold by decreasing the concentration of intermediate metabolites and reducing flux through competing pathways. Inducible compartmentalization of enzymes into synthetic organelles can thus be used to control engineered metabolic pathways, limit intermediates and favor the formation of desired products.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All plasmids, strains and raw data will be made available upon reasonable request to the corresponding authors.
References
Nielsen, J. & Keasling, J. D. Engineering cellular metabolism. Cell 164, 1185–1197 (2016).
Keasling, J. D. Manufacturing molecules through metabolic engineering. Science 50, 1355 (2011).
Ajikumar, P. K. et al. Isoprenoid pathway optimization for Taxol precursor overproduction in Escherichia coli. Science 330, 70–74 (2010).
Lalwani, M. A., Zhao, E. M. & Avalos, J. L. Current and future modalities of dynamic control in metabolic engineering. Curr. Opin. Biotechnol. 52, 56–65 (2018).
Tomala, K. & Korona, R. Evaluating the fitness cost of protein expression in Saccharomyces cerevisiae. Genome Biol. Evol. 5, 2051–2060 (2013).
Brockman, I. M. & Prather, K. L. J. Dynamic knockdown of E. coli central metabolism for redirecting fluxes of primary metabolites. Metab. Eng. 28, 104–113 (2015).
Tan, S. Z. & Prather, K. L. Dynamic pathway regulation: recent advances and methods of construction. Curr. Opin. Chem. Biol. 41, 28–35 (2017).
Thomik, T., Wittig, I., Choe, J. Y., Boles, E. & Oreb, M. An artificial transport metabolon facilitates improved substrate utilization in yeast. Nat. Chem. Biol. 13, 1158–1163 (2017).
Lin, J. L., Zhu, J. & Wheeldon, I. Synthetic protein scaffolds for biosynthetic pathway colocalization on lipid droplet membranes. ACS Synth. Biol. 6, 1534–1544 (2017).
Pedley, A. M. & Benkovic, S. J. A new view into the regulation of purine metabolism: the purinosome. Trends Biochem. Sci. 42, 141–154 (2017).
French, J. B. et al. Spatial colocalization and functional link of purinosomes with mitochondria. Science 351, 733–737 (2016).
Zhang, Y. et al. Protein–protein interactions and metabolite channelling in the plant tricarboxylic acid cycle. Nat. Commun. 8, 15212 (2017).
Narayanaswamy, R. et al. Widespread reorganization of metabolic enzymes into reversible assemblies upon nutrient starvation. Proc. Natl Acad. Sci. USA 106, 10147–10152 (2009).
Kistler, H. C. & Broz, K. Cellular compartmentalization of secondary metabolism. Front. Microbiol. 6, 1–11 (2015).
Castellana, M. et al. Enzyme clustering accelerates processing of intermediates through metabolic channeling. Nat. Biotechnol. 32, 1011–1018 (2014).
George, K. W. et al. Integrated analysis of isopentenyl pyrophosphate (IPP) toxicity in isoprenoid-producing Escherichia coli. Metab. Eng. 47, 60–72 (2018).
Dueber, J. E. et al. Synthetic protein scaffolds provide modular control over metabolic flux. Nat. Biotechnol. 27, 753–759 (2009).
Li, T., Chen, X., Cai, Y. & Dai, J. Artificial Protein Scaffold System (AProSS): an efficient method to optimize exogenous metabolic pathways in Saccharomyces cerevisiae. Metab. Eng. 49, 13–20 (2018).
Lau, Y. H., Giessen, T. W., Altenburg, W. J. & Silver, P. A. Prokaryotic nanocompartments form synthetic organelles in a eukaryote. Nat. Commun. 9, 1311 (2018).
Avalos, J. L., Fink, G. R. & Stephanopoulos, G. Compartmentalization of metabolic pathways in yeast mitochondria improves the production of branched-chain alcohols. Nat. Biotechnol. 31, 335–341 (2013).
DeLoache, W. C., Russ, Z. N. & Dueber, J. E. Towards repurposing the yeast peroxisome for compartmentalizing heterologous metabolic pathways. Nat. Commun. 7, 11152 (2016).
Hammer, S. K. & Avalos, J. L. Harnessing yeast organelles for metabolic engineering. Nat. Chem. Biol. 13, 823–832 (2017).
Toettcher, J. E., Voigt, Ca, Weiner, O. D. & Lim, Wa The promise of optogenetics in cell biology: interrogating molecular circuits in space and time. Nat. Methods 8, 35–38 (2011).
Shin, Y. et al. Spatiotemporal control of intracellular phase transitions using light-activated optoDroplets. Cell 168, 159–171.e14 (2017).
Dine, E., Gil, A. A., Uribe, G., Brangwynne, C. P. & Toettcher, J. E. Protein phase separation provides long-term memory of transient spatial stimuli. Cell Syst. 6, 655–663 (2018).
Taslimi, A. et al. An optimized optogenetic clustering tool for probing protein interaction and function. Nat. Commun. 5, 4925 (2014).
Nakamura, H. et al. Intracellular production of hydrogels and synthetic RNA granules by multivalent molecular interactions. Nat. Mater. 17, 79–88 (2018).
Bugaj, L. J., Choksi, A. T., Mesuda, C. K., Kane, R. S. & Schaffer, D. V. Optogenetic protein clustering and signaling activation in mammalian cells. Nat. Methods 10, 249–252 (2013).
Gil, A. A. et al. Photoactivation of the BLUF protein PixD probed by the site-specific incorporation of fluorotyrosine residues. J. Am. Chem. Soc. 139, 14638–14648 (2017).
Shemiakina, I. I. et al. A monomeric red fluorescent protein with low cytotoxicity. Nat. Commun. 3, 1204 (2012).
Shin, Y. & Brangwynne, C. P. Liquid phase condensation in cell physiology and disease. Science 357, eaaf4382 (2017).
Yuan, J. & Ching, C. B. Combinatorial assembly of large biochemical pathways into yeast chromosomes for improved production of value-added compounds. ACS Synth. Biol. 4, 23–31 (2014).
Zhao, E. M. et al. Optogenetic regulation of engineered cellular metabolism for microbial chemical production. Nature 555, 683–687 (2018).
Yuan, H. & Bauer, C. E. PixE promotes dark oligomerization of the BLUF photoreceptor PixD. Proc. Natl Acad. Sci. USA 105, 11715–11719 (2008).
Brachmann, C. B. et al. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14, 115–132 (1998).
Giaever, G. & Nislow, C. The yeast deletion collection: a decade of functional genomics. Genetics 197, 451–465 (2014).
Entian, K. D. & Kötter, P. 25 Yeast genetic strain and plasmid collections. Meth. Microbiol. 36, 629–666 (2007).
Lee, M. E., Aswani, A., Han, A. S., Tomlin, C. J. & Dueber, J. E. Expression-level optimization of a multi-enzyme pathway in the absence of a high-throughput assay. Nucleic Acids Res. 41, 10668–10678 (2013).
Bracha, D. et al. Mapping local and global liquid phase behavior in living cells using photo-oligomerizable seeds. Cell 175, 1467–1480.e13 (2018).
Ryan, K. S., Balibar, C. J., Turo, K. E., Walsh, C. T. & Drennan, C. L. The violacein biosynthetic enzyme VioE shares a fold with lipoprotein transporter proteins. J. Biol. Chem. 283, 6467–6475 (2008).
Martin, V. J. J., Pitera, D. J., Withers, S. T., Newman, J. D. & Keasling, J. D. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nature 21, 796–802 (2003).
Ro, D. et al. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature 440, 3–6 (2006).
Hua Yuan, H. et al. Mutational and Structural Studies of the PixD BLUF Output Signal That Affects Light-Regulated Interactions with PixE. Biochemistry 50, 6365–6375 (2011).
Acknowledgements
We thank all members of the Toettcher and Avalos laboratories for helpful comments. We also thank J. Dueber for kindly providing violacein enzyme plasmids. This work was supported by the Maeder Graduate Fellowship in Energy and the Environment (to E.M.Z.), NIH grant DP2EB024247 (to J.E.T.) and The Pew Charitable Trusts, the U.S. DOE Office of Biological and Environmental Research, Genomic Science Program Award DESC0019363, and NSF CAREER Award CBET-1751840 (to J.L.A.) and a Schmidt Transformative Technology grant (to J.E.T. and J.L.A).
Author information
Authors and Affiliations
Contributions
E.M.Z., M.Z.W., J.E.T. and J.L.A. conceived the project and designed the experiments. E.M.Z. and N.S. conducted all metabolic flux experiments. E.M.Z., N.S., M.Z.W., E.D. and N.L.P. cloned constructs and performed microscopy. Z.G. contributed methodology and reagents. E.M.Z., J.E.T. and J.L.A. wrote the paper with editing from all authors. J.E.T. and J.L.A. provided funding and supervised the research.
Corresponding authors
Ethics declarations
Competing interests
Some of the authors are co-inventors on patent applications harnessing optogenetics for metabolic engineering (J.L.A., J.E.T. and E.M.Z.: patent application no. WO2017177147A1) and establishing optogenetic control of protein clustering (J.E.T.: patent application no. US20170355977A1).
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Tables 1 and 2, Supplementary Figures 1–11, Supplementary Note 1
Supplementary Video 1
OptoDroplet formation and dissociation in S. cerevisiae.
Supplementary Video 2
OptoCluster formation and dissociation in S. cerevisiae.
Supplementary Video 3
PixELL formation and dissociation in S. cerevisiae.
Rights and permissions
About this article
Cite this article
Zhao, E.M., Suek, N., Wilson, M.Z. et al. Light-based control of metabolic flux through assembly of synthetic organelles. Nat Chem Biol 15, 589–597 (2019). https://doi.org/10.1038/s41589-019-0284-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-019-0284-8
This article is cited by
-
Optogenetic control of mRNA condensation reveals an intimate link between condensate material properties and functions
Nature Communications (2024)
-
Combinatorial optimization of gene expression through recombinase-mediated promoter and terminator shuffling in yeast
Nature Communications (2024)
-
Technologies for studying phase-separated biomolecular condensates
Advanced Biotechnology (2024)
-
Microbial cell factories based on filamentous bacteria, yeasts, and fungi
Microbial Cell Factories (2023)
-
RNAs undergo phase transitions with lower critical solution temperatures
Nature Chemistry (2023)