Abstract
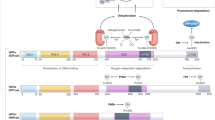
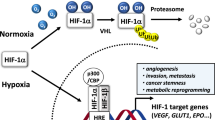
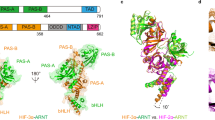
Hypoxia-inducible factor-2 (HIF-2) is a heterodimeric transcription factor formed through dimerization between an oxygen-sensitive HIF-2α subunit and its obligate partner subunit ARNT. Enhanced HIF-2 activity drives some cancers, whereas reduced activity causes anemia in chronic kidney disease. Therefore, modulation of HIF-2 activity via direct-binding ligands could provide many new therapeutic benefits. Here, we explored HIF-2α chemical ligands using combined crystallographic, biophysical, and cell-based functional studies. We found chemically unrelated antagonists to employ the same mechanism of action. Their binding displaced residue M252 from inside the HIF-2α PAS-B pocket toward the ARNT subunit to weaken heterodimerization. We also identified first-in-class HIF-2α agonists and found that they significantly displaced pocket residue Y281. Its dramatic side chain movement increases heterodimerization stability and transcriptional activity. Our findings show that despite binding to the same HIF-2α PAS-B pocket, ligands can manifest as inhibitors versus activators by mobilizing different pocket residues to allosterically alter HIF-2α–ARNT heterodimerization.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
References
Wu, D. & Rastinejad, F. Structural characterization of mammalian bHLH-PAS transcription factors. Curr. Opin. Struct. Biol. 43, 1–9 (2017).
Möglich, A., Ayers, R. A. & Moffat, K. Structure and signaling mechanism of Per-ARNT-Sim domains. Structure 17, 1282–1294 (2009).
Wu, D., Su, X., Potluri, N., Kim, Y. & Rastinejad, F. NPAS1-ARNT and NPAS3-ARNT crystal structures implicate the bHLH-PAS family as multi-ligand binding transcription factors. eLife 5, e18790 (2016).
McIntosh, B. E., Hogenesch, J. B. & Bradfield, C. A. Mammalian Per-Arnt-Sim proteins in environmental adaptation. Annu. Rev. Physiol. 72, 625–645 (2010).
Schito, L. & Semenza, G. L. Hypoxia-inducible factors: master regulators of cancer progression. Trends Cancer 2, 758–770 (2016).
Keith, B., Johnson, R. S. & Simon, M. C. HIF1α and HIF2α: sibling rivalry in hypoxic tumour growth and progression. Nat. Rev. Cancer 12, 9–22 (2011).
Ravenna, L., Salvatori, L. & Russo, M. A. HIF3α: the little we know. FEBS. J. 283, 993–1003 (2016).
Wu, D., Potluri, N., Lu, J., Kim, Y. & Rastinejad, F. Structural integration in hypoxia-inducible factors. Nature 524, 303–308 (2015).
Ivan, M. et al. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292, 464–468 (2001).
Jaakkola, P. et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292, 468–472 (2001).
Yu, F., WhiteS. B., Zhao, Q. & Lee, F. S. HIF-1α binding to VHL is regulated by stimulus-sensitive proline hydroxylation. Proc. Natl Acad. Sci. USA 98, 9630–9635 (2001).
Lando, D., Peet, D. J., Whelan, D. A., Gorman, J. J. & Whitelaw, M. L. Asparagine hydroxylation of the HIF transactivation domain a hypoxic switch. Science 295, 858–861 (2002).
Lando, D. et al. FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev. 16, 1466–1471 (2002).
Huang, P., Chandra, V. & Rastinejad, F. Structural overview of the nuclear receptor superfamily: insights into physiology and therapeutics. Annu. Rev. Physiol. 72, 247–272 (2010).
Denison, M. S. & Nagy, S. R. Activation of the aryl hydrocarbon receptor by structurally diverse exogenous and endogenous chemicals. Annu. Rev. Pharmacol. Toxicol. 43, 309–334 (2003).
Denison, M. S., Soshilov, A. A., He, G., DeGroot, D. E. & Zhao, B. Exactly the same but different: promiscuity and diversity in the molecular mechanisms of action of the aryl hydrocarbon (dioxin) receptor. Toxicol. Sci. 124, 1–22 (2011).
Scheuermann, T. H. et al. Artificial ligand binding within the HIF2α PAS-B domain of the HIF2 transcription factor. Proc. Natl Acad. Sci. USA 106, 450–455 (2009).
Key, J., Scheuermann, T. H., Anderson, P. C., Daggett, V. & Gardner, K. H. Principles of ligand binding within a completely buried cavity in HIF2αa PAS-B. J. Am. Chem. Soc. 131, 17647–17654 (2009).
Cardoso, R. et al. Identification of Cys255 in HIF-1α as a novel site for development of covalent inhibitors of HIF-1α/ARNT PasB domain protein-protein interaction. Protein Sci. 21, 1885–1896 (2012).
Guo, Y. et al. Regulating the ARNT/TACC3 axis: multiple approaches to manipulating protein/protein interactions with small molecules. ACS. Chem. Biol. 8, 626–635 (2013).
Fala, A. M. et al. Unsaturated fatty acids as high-affinity ligands of the C-terminal Per-ARNT-Sim domain from the hypoxia-inducible factor 3α. Sci. Rep. 5, 12698 (2015).
Hewitson, K. S. & Schofield, C. J. The HIF pathway as a therapeutic target. Drug Discov. Today 9, 704–711 (2004).
Wallace, E. M. et al. A small-molecule antagonist of HIF2α is efficacious in preclinical models of renal cell carcinoma. Cancer Res. 76, 5491–5500 (2016).
Chen, W. et al. Targeting renal cell carcinoma with a HIF-2 antagonist. Nature 539, 112–117 (2016).
Cho, H. et al. On-target efficacy of a HIF-2α antagonist in preclinical kidney cancer models. Nature 539, 107–111 (2016).
Maxwell, P. H. & Eckardt, K. U. HIF prolyl hydroxylase inhibitors for the treatment of renal anaemia and beyond. Nat. Rev. Nephrol. 12, 157–168 (2016).
Yousaf, F. & Spinowitz, B. Hypoxia-inducible factor stabilizers: a new avenue for reducing BP while helping hemoglobin? Curr. Hypertens. Rep. 18, 23 (2016).
Gupta, N. & Wish, J. B. Hypoxia-inducible factor prolyl hydroxylase inhibitors: a potential new treatment for anemia in patients With CKD. Am. J. Kidney. Dis. 69, 815–826 (2017).
Yeh, T. L. et al. Molecular and cellular mechanisms of HIF prolyl hydroxylase inhibitors in clinical trials. Chem. Sci. 8, 7651–7668 (2017).
Seidel, S. A. et al. Microscale thermophoresis quantifies biomolecular interactions under previously challenging conditions. Methods 59, 301–315 (2013).
Ciulli, A. & Abell, C. Fragment-based approaches to enzyme inhibition. Curr. Opin. Biotechnol. 18, 489–496 (2007).
Scheuermann, T. H. et al. Allosteric inhibition of hypoxia inducible factor-2 with small molecules. Nat. Chem. Biol. 9, 271–276 (2013).
Forbes, S. A. et al. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 43, D805–D811 (2015).
Annis, D. A., Nickbarg, E., Yang, X., Ziebell, M. R. & Whitehurst, C. E. Affinity selection-mass spectrometry screening techniques for small molecule drug discovery. Curr. Opin. Chem. Biol. 11, 518–526 (2007).
Bonomini, M., Del Vecchio, L., Sirolli, V. & Locatelli, F. New treatment approaches for the anemia of CKD. Am. J. Kidney. Dis. 67, 133–142 (2016).
Besarab, A. et al. Roxadustat (FG-4592): correction of anemia in incident dialysis patients. J. Am. Soc. Nephrol. 27, 1225–1233 (2016).
Brigandi, R. A. et al. A novel hypoxia-inducible factor-prolyl hydroxylase inhibitor (GSK1278863) for anemia in CKD: a 28-day, phase 2A randomized trial. Am. J. Kidney. Dis. 67, 861–871 (2016).
Pergola, P. E., Spinowitz, B. S., Hartman, C. S., Maroni, B. J. & Haase, V. H. Vadadustat, a novel oral HIF stabilizer, provides effective anemia treatment in nondialysis-dependent chronic kidney disease. Kidney Int. 90, 1115–1122 (2016).
Beck, H. et al. Discovery of molidustat (BAY 85-3934): a small-molecule oral HIF-prolyl hydroxylase (HIF-PH) inhibitor for the treatment of renal anemia. ChemMedChem 13, 988–1003 (2018).
Rogers, J. L. et al. Development of inhibitors of the PAS-B domain of the HIF-2α transcription factor. J. Med. Chem. 56, 1739–1747 (2013).
Scheuermann, T. H. et al. Isoform-selective and stereoselective inhibition of hypoxia inducible factor-2. J. Med. Chem. 58, 5930–5941 (2015).
Wehn, P. M. et al. Design and activity of specific hypoxia-inducible factor-2α (HIF-2α) inhibitors for the treatment of clear cell renal cell carcinoma: discovery of clinical candidate (S)-3-((2,2-difluoro-1-hydroxy-7-(methylsulfonyl)-2,3-dihydro-1H-inden-4-yl)oxy)-5-fluorobenzonitrile (PT2385). J. Med. Chem. 61, 9691–9721 (2018).
Minor, W., Cymborowski, M., Otwinowski, Z. & Chruszcz, M. HKL-3000: the integration of data reduction and structure solution--from diffraction images to an initial model in minutes. Acta Crystallogr. D. Biol. Crystallogr. 62, 859–866 (2006).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 66, 486–501 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D. Biol. Crystallogr. 66, 12–21 (2010).
Marsh, J. J. et al. Structural insights into fibrinogen dynamics using amide hydrogen/deuterium exchange mass spectrometry. Biochemistry 52, 5491–5502 (2013).
Woods, V. L. Jr. & Hamuro, Y. High resolution, high-throughput amide deuterium exchange-mass spectrometry (DXMS) determination of protein binding site structure and dynamics: utility in pharmaceutical design. J. Cell. Biochem. Suppl. 84, 89–98 (2001).
Walters, B. T., Ricciuti, A., Mayne, L. & Englander, S. W. Minimizing back exchange in the hydrogen exchange-mass spectrometry experiment. J. Am. Soc. Mass. Spectrom. 23, 2132–2139 (2012).
Li, S. et al. Mechanism of intracellular cAMP sensor Epac2 activation: cAMP-induced conformational changes identified by amide hydrogen/deuterium exchange mass spectrometry (DXMS). J. Biol. Chem. 286, 17889–17897 (2011).
Acknowledgements
We thank members of the Structural Biology Center at Argonne National Laboratory for their help with data collection at the 19-ID beamline, D. Liu and I. Pass at Sanford Burnham Prebys for kindly providing materials, and G.N. Murshudov at University of Cambridge for the help with ligand building at the 2016 CCP4/APS summer school. This work was supported by the Wellcome Trust and by grants from the National Institutes of Health (R01GM117013, R01DK118297) and US ARMY Medical Research (W81XWH-16-1-0322) to F.R., as well as grants from Shandong University (Qilu Young Scholar 86963072), National Natural Science Foundation of China (31700114), Natural Science Foundation of Jiangsu Province (BK20170399), and the 111 Project (B16030) to D.W.
Author information
Authors and Affiliations
Contributions
D.W. purified proteins, carried out crystallization and solved the structures. X.S. conducted cell-based experiments. J.L. purified proteins and performed biochemical assays. S.L. executed the H/D-ex MS analysis. B.L.H. and S.V. performed the TR-FRET binding assays and thermal-shift-based screening experiments. N.P. produced the expression and mutation constructs. X.D. contributed to biochemical assays and structure refinement. Y.K. collected and processed synchrotron diffraction data. S.K. provided critical instruments and training for biochemical studies. All authors analyzed results. D.W. and F.R. conceived this study, designed experiments and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1 and 2, Supplementary Figures 1–14
Rights and permissions
About this article
Cite this article
Wu, D., Su, X., Lu, J. et al. Bidirectional modulation of HIF-2 activity through chemical ligands. Nat Chem Biol 15, 367–376 (2019). https://doi.org/10.1038/s41589-019-0234-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-019-0234-5
This article is cited by
-
Targeting hypoxia-inducible factors: therapeutic opportunities and challenges
Nature Reviews Drug Discovery (2024)
-
Hypoxia-induced signaling in the cardiovascular system: pathogenesis and therapeutic targets
Signal Transduction and Targeted Therapy (2023)
-
Expression and Clinical Significance of HIF-1α in Follicular Fluid and Granulosa Cells in Infertile PCOS Patients
Reproductive Sciences (2023)
-
Identification of oleoylethanolamide as an endogenous ligand for HIF-3α
Nature Communications (2022)
-
Structural insight into the ligand binding mechanism of aryl hydrocarbon receptor
Nature Communications (2022)