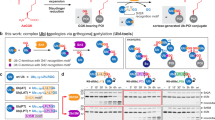
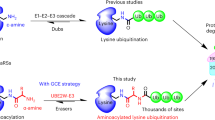
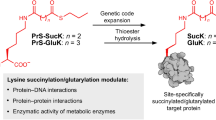
Abstract
Post-translational modification of proteins with ubiquitin and ubiquitin-like proteins (Ubls) is central to the regulation of eukaryotic cellular processes. Our ability to study the effects of ubiquitylation, however, is limited by the difficulty to prepare homogenously modified proteins in vitro and by the impossibility to selectively trigger specific ubiquitylation events in living cells. Here we combine genetic-code expansion, bioorthogonal Staudinger reduction and sortase-mediated transpeptidation to develop a general tool to ubiquitylate proteins in an inducible fashion. The generated ubiquitin conjugates display a native isopeptide bond and bear two point mutations in the ubiquitin C terminus that confer resistance toward deubiquitinases. Nevertheless, physiological integrity of sortase-generated diubiquitins in decoding cellular functions via recognition by ubiquitin-binding domains is retained. Our approach allows the site-specific attachment of Ubls to nonrefoldable, multidomain proteins and enables inducible and ubiquitin-ligase-independent ubiquitylation of proteins in mammalian cells, providing a powerful tool to dissect the biological functions of ubiquitylation with temporal control.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The authors declare that the data supporting the findings of this study are available in the paper and its supplementary information files. Raw data and other materials are available upon reasonable request.
References
Hershko, A. & Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 (1998).
Komander, D. & Rape, M. The ubiquitin code. Annu. Rev. Biochem. 81, 203–229 (2012).
Kulathu, Y. & Komander, D. Atypical ubiquitylation - the unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages. Nat. Rev. Mol. Cell Biol. 13, 508–523 (2012).
Komander, D., Clague, M. J. & Urbé, S. Breaking the chains: structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 10, 550–563 (2009).
van der Veen, A. G. & Ploegh, H. L. Ubiquitin-like proteins. Annu. Rev. Biochem. 81, 323–357 (2012).
Mali, S. M., Singh, S. K., Eid, E. & Brik, A. Ubiquitin signaling: chemistry comes to the rescue. J. Am. Chem. Soc. 139, 4971–4986 (2017).
Trang, V. H. et al. Nonenzymatic polymerization of ubiquitin: single-step synthesis and isolation of discrete ubiquitin oligomers. Angew. Chem. Int. Ed. Engl. 51, 13085–13088 (2012).
Chen, J., Ai, Y., Wang, J., Haracska, L. & Zhuang, Z. Chemically ubiquitylated PCNA as a probe for eukaryotic translesion DNA synthesis. Nat. Chem. Biol. 6, 270–272 (2010).
Weikart, N. D. & Mootz, H. D. Generation of site-specific and enzymatically stable conjugates of recombinant proteins with ubiquitin-like modifiers by the Cu(i)-catalyzed azide-alkyne cycloaddition. Chembiochem 11, 774–777 (2010).
Eger, S., Scheffner, M., Marx, A. & Rubini, M. Synthesis of defined ubiquitin dimers. J. Am. Chem. Soc. 132, 16337–16339 (2010).
Stanley, M. & Virdee, S. Genetically directed production of recombinant, isosteric and nonhydrolysable ubiquitin conjugates. Chembiochem 17, 1472–1480 (2016).
Virdee, S., Ye, Y., Nguyen, D. P., Komander, D. & Chin, J. W. Engineered diubiquitin synthesis reveals Lys29-isopeptide specificity of an OTU deubiquitinase. Nat. Chem. Biol. 6, 750–757 (2010).
Li, X., Fekner, T., Ottesen, J. J. & Chan, M. K. A pyrrolysine analogue for site-specific protein ubiquitination. Angew. Chem. Int. Ed. Engl. 48, 9184–9187 (2009).
Virdee, S. et al. Traceless and site-specific ubiquitination of recombinant proteins. J. Am. Chem. Soc. 133, 10708–10711 (2011).
Stanley, M. & Virdee, S. Chemical ubiquitination for decrypting a cellular code. Biochem. J. 473, 1297–1314 (2016).
Spasser, L. & Brik, A. Chemistry and biology of the ubiquitin signal. Angew. Chem. Int. Ed. Engl. 51, 6840–6862 (2012).
Pham, G. H. & Strieter, E. R. Peeling away the layers of ubiquitin signaling complexities with synthetic ubiquitin-protein conjugates. Curr. Opin. Chem. Biol. 28, 57–65 (2015).
Mazmanian, S. K., Liu, G., Ton-That, H. & Schneewind, O. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science 285, 760–763 (1999).
Popp, M. W. & Ploegh, H. L. Making and breaking peptide bonds: protein engineering using sortase. Angew. Chem. Int. Ed. Engl. 50, 5024–5032 (2011).
Chen, I., Dorr, B. M. & Liu, D. R. A general strategy for the evolution of bond-forming enzymes using yeast display. Proc. Natl Acad. Sci. USA 108, 11399–11404 (2011).
Luo, J., Liu, Q., Morihiro, K. & Deiters, A. Small-molecule control of protein function through Staudinger reduction. Nat. Chem. 8, 1027–1034 (2016).
Lang, K. & Chin, J. W. Cellular incorporation of unnatural amino acids and bioorthogonal labeling of proteins. Chem. Rev. 114, 4764–4806 (2014).
Liu, C. C. & Schultz, P. G. Adding new chemistries to the genetic code. Annu. Rev. Biochem. 79, 413–444 (2010).
Hancock, S. M., Uprety, R., Deiters, A. & Chin, J. W. Expanding the genetic code of yeast for incorporation of diverse unnatural amino acids via a pyrrolysyl-tRNA synthetase/tRNA pair. J. Am. Chem. Soc. 132, 14819–14824 (2010).
Lang, K. et al. Genetically encoded norbornene directs site-specific cellular protein labelling via a rapid bioorthogonal reaction. Nat. Chem. 4, 298–304 (2012).
Li, F. et al. Expanding the genetic code for photoclick chemistry in E. coli, mammalian cells, and A. thaliana. Angew. Chem. Int. Ed. Engl. 52, 9700–9704 (2013).
Greiss, S. & Chin, J. W. Expanding the genetic code of an animal. J. Am. Chem. Soc. 133, 14196–14199 (2011).
Bianco, A., Townsley, F. M., Greiss, S., Lang, K. & Chin, J. W. Expanding the genetic code of Drosophila melanogaster. Nat. Chem. Biol. 8, 748–750 (2012).
Han, S. et al. Expanding the genetic code of Mus musculus. Nat. Commun. 8, 14568 (2017).
Wang, L., Brock, A., Herberich, B. & Schultz, P. G. Expanding the genetic code of Escherichia coli. Science 292, 498–500 (2001).
Neumann, H., Peak-Chew, S. Y. & Chin, J. W. Genetically encoding N(ε)-acetyllysine in recombinant proteins. Nat. Chem. Biol. 4, 232–234 (2008).
Dorr, B. M., Ham, H. O., An, C., Chaikof, E. L. & Liu, D. R. Reprogramming the specificity of sortase enzymes. Proc. Natl Acad. Sci. USA 111, 13343–13348 (2014).
Békés, M. et al. DUB-resistant ubiquitin to survey ubiquitination switches in mammalian cells. Cell Rep. 5, 826–838 (2013).
Husnjak, K. & Dikic, I. Ubiquitin-binding proteins: decoders of ubiquitin-mediated cellular functions. Annu. Rev. Biochem. 81, 291–322 (2012).
Kulathu, Y., Akutsu, M., Bremm, A., Hofmann, K. & Komander, D. Two-sided ubiquitin binding explains specificity of the TAB2 NZF domain. Nat. Struct. Mol. Biol. 16, 1328–1330 (2009).
Zhang, X. et al. An interaction landscape of ubiquitin signaling. Mol. Cell 65, 941–955.e8 (2017).
Varadan, R., Assfalg, M., Raasi, S., Pickart, C. & Fushman, D. Structural determinants for selective recognition of a Lys48-linked polyubiquitin chain by a UBA domain. Mol. Cell 18, 687–698 (2005).
Sims, J. J. & Cohen, R. E. Linkage-specific avidity defines the lysine 63-linked polyubiquitin-binding preference ofrap80. Mol. Cell 33, 775–783 (2009).
Sato, Y. et al. Structural basis for specific recognition of Lys 63-linked polyubiquitin chains by tandem UIMs of RAP80. EMBO J. 28, 2461–2468 (2009).
Bravo, R., Frank, R., Blundell, P. A. & Macdonald-Bravo, H. Cyclin/PCNA is the auxiliary protein of DNA polymerase-delta. Nature 326, 515–517 (1987).
Hoege, C., Pfander, B., Moldovan, G. L., Pyrowolakis, G. & Jentsch, S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419, 135–141 (2002).
Flotho, A. & Melchior, F. Sumoylation: a regulatory protein modification in health and disease. Annu. Rev. Biochem. 82, 357–385 (2013).
Pawale, V. S., Yadav, P. & Roy, R. P. Facile one-step assembly of bona fide SUMO conjugates by chemoenzymatic ligation. Chembiochem 19, 1137–1141 (2018).
Suree, N. et al. The structure of the Staphylococcus aureus sortase-substrate complex reveals how the universally conserved LPXTG sorting signal is recognized. J. Biol. Chem. 284, 24465–24477 (2009).
Hirakawa, H., Ishikawa, S. & Nagamune, T. Design of Ca2+-independent Staphylococcus aureus sortase A mutants. Biotechnol. Bioeng. 109, 2955–2961 (2012).
Keren-Kaplan, T. et al. Synthetic biology approach to reconstituting the ubiquitylation cascade in bacteria. EMBO J. 31, 378–390 (2012).
Martinez-Fonts, K. & Matouschek, A. A rapid and versatile method for generating proteins with defined ubiquitin chains. Biochemistry 55, 1898–1908 (2016).
Nguyen, T. A., Cigler, M. & Lang, K. Expanding the genetic code to study protein-protein interactions. Angew. Chem. Int. Ed. Egnl. 57, 14350–14361 (2018).
Cigler, M. et al. Proximity-triggered covalent stabilization of low-affinity protein complexes in vitro and in vivo. Angew. Chem. Int. Ed. Engl. 56, 15737–15741 (2017).
David, Y., Vila-Perelló, M., Verma, S. & Muir, T. W. Chemical tagging and customizing of cellular chromatin states using ultrafast trans-splicing inteins. Nat. Chem. 7, 394–402 (2015).
Blizzard, R. J. et al. Ideal bioorthogonal reactions using a site-specifically encoded tetrazine amino acid. J. Am. Chem. Soc. 137, 10044–10047 (2015).
Pickart, C. M. & Raasi, S. Ubiquitin and protein degradation, part B: controlled synthesis of polyubiquitin chains. in Methods in Enzymology, Vol. 399 (ed. Deshaies, R. J.) 26–28 (Elsevier, 2005).
Berndsen, C. E. & Wolberger, C. A spectrophotometric assay for conjugation of ubiquitin and ubiquitin-like proteins. Anal. Biochem. 418, 102–110 (2011).
Bremm, A., Freund, S. M. & Komander, D. Lys11-linked ubiquitin chains adopt compact conformations and are preferentially hydrolyzed by the deubiquitinase Cezanne. Nat. Struct. Mol. Biol. 17, 939–947 (2010).
Komander, D. et al. Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. EMBO Rep. 10, 466–473 (2009).
Best, R. B. et al. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ(1) and χ(2) dihedral angles. J. Chem. Theory. Comput. 8, 3257–3273 (2012).
Abraham, M. J. et al. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2, 19–25 (2015).
Humphrey, W., Dalke, A. & Schulten, K. VMD: visual molecular dynamics. J. Mol. Graph. 14, 27–28 (1996).
Schmied, W. H., Elsässer, S. J., Uttamapinant, C. & Chin, J. W. Efficient multisite unnatural amino acid incorporation in mammalian cells via optimized pyrrolysyl tRNA synthetase/tRNA expression and engineered eRF1. J. Am. Chem. Soc. 136, 15577–15583 (2014).
Acknowledgements
This work was supported by the Excellence Initiative CIPSM and the DFG through the following programmes: GRK1721, SFB1309 and SPP1623 (to K.L.) as well as SFB1035 project B12 to V.R.I.K. and B10 to K.L. K.L. is a Mössbauer Professor at TUM-IAS. We thank C. Biertümpfel, MPI Martinsried for PCNA plasmids. Natively ubiquitylated PCNA was a generous gift from C. Biertümpfel. We thank M. Vermeulen, Radboud Insitute for Molecular Life sciences for GST-TAB2-NZF plasmid.
Author information
Authors and Affiliations
Contributions
K.L. conceived the research plan and experimental strategy. M.F. synthesized UAAs, performed all experiments in bacteria, including cloning, expression, purification of proteins and ubiquitylation/SUMOylation assays, as well as enzymatic assays and pull-down assays. A.-D.B. created PylRS libraries and evolved AzGGKRS. D.H.-G. performed initial mammalian cell experiments, including site-specific incorporation of AzGGK into HEK293T cells, and V.B. and A.B. performed ubiquitylation and SUMOylation assays in live HEK293T cells. A.J. and V.R.I.K. performed MD simulations. All authors analyzed data, and K.L. wrote the paper with input from the other authors.
Corresponding author
Ethics declarations
Competing interests
We have filed a patent concerning the sortylation approach.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1–9, Supplementary Figures 1–23, Supplementary Notes 1 and 2
Rights and permissions
About this article
Cite this article
Fottner, M., Brunner, AD., Bittl, V. et al. Site-specific ubiquitylation and SUMOylation using genetic-code expansion and sortase. Nat Chem Biol 15, 276–284 (2019). https://doi.org/10.1038/s41589-019-0227-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-019-0227-4
This article is cited by
-
Structure-guided engineering enables E3 ligase-free and versatile protein ubiquitination via UBE2E1
Nature Communications (2024)
-
Recent advances in chemical protein synthesis: method developments and biological applications
Science China Chemistry (2024)
-
Nature-inspired protein ligation and its applications
Nature Reviews Chemistry (2023)
-
A new dawn beyond lysine ubiquitination
Nature Chemical Biology (2022)
-
Tuning flavin environment to detect and control light-induced conformational switching in Drosophila cryptochrome
Communications Biology (2021)