Abstract
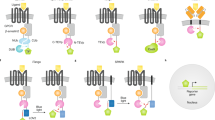
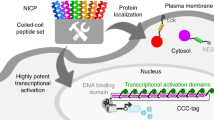
Cellular signal transduction is predominantly based on protein interactions and their post-translational modifications, which enable a fast response to input signals. Owing to difficulties in designing new unique protein–protein interactions, designed cellular logic has focused on transcriptional regulation; however, that process has a substantially slower response, because it requires transcription and translation. Here, we present de novo design of modular, scalable signaling pathways based on proteolysis and designed coiled coils (CC) and implemented in mammalian cells. A set of split proteases with highly specific orthogonal cleavage motifs was constructed and combined with strategically positioned cleavage sites and designed orthogonal CC dimerizing domains with tunable affinity for competitive displacement after proteolytic cleavage. This framework enabled the implementation of Boolean logic functions and signaling cascades in mammalian cells. The designed split-protease-cleavable orthogonal-CC-based (SPOC) logic circuits enable response to chemical or biological signals within minutes rather than hours and should be useful for diverse medical and nonmedical applications.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The authors declare that the data supporting the findings of this study are available in the paper and its supplementary information files. A set of plasmids for SPOC logic, comprising the split orthogonal proteases, cycLuc reporters and CC-building modules, have been deposited with Addgene under Addgene IDs 118966, 118967, 118968, 118969, 118970, 119182, 119207, 119208, 119209, 119210, 119211, 119212, 119213, 119214, 119299, 119300, 119302 and 119303. The raw data are available from the corresponding author upon reasonable request.
References
Cohen, G. B., Ren, R. & Baltimore, D. Modular binding domains in signal transduction proteins. Cell 80, 237–248 (1995).
Nandagopal, N. & Elowitz, M. B. Synthetic biology: integrated gene circuits. Science 333, 1244–1248 (2011).
Good, M. C., Zalatan, J. G. & Lim, W. A. Scaffold proteins: hubs for controlling the flow of cellular information. Science 332, 680–686 (2011).
Gordley, R. M. et al. Engineering dynamical control of cell fate switching using synthetic phospho-regulons. Proc. Natl Acad. Sci. USA 113, 13528–13533 (2016).
Dueber, J. E., Yeh, B. J., Chak, K. & Lim, W. A. Reprogramming control of an allosteric signaling switch through modular recombination. Science 301, 1904–1908 (2003).
Howard, P. L., Chia, M. C., Del Rizzo, S., Liu, F.-F. & Pawson, T. Redirecting tyrosine kinase signaling to an apoptotic caspase pathway through chimeric adaptor proteins. Proc. Natl Acad. Sci. USA 100, 11267–11272 (2003).
Lonzaric, J., Fink, T. & Jerala, R. Design and applications of synthetic information processing circuits in mammalian cells. in Synthetic Biology Vol. 2, 1–34 (Royal Society of Chemistry, London, 2018).
Haellman, V. & Fussenegger, M. Synthetic biology: engineering cell-based biomedical devices. Curr. Opin. Biomed. Eng. 4, 50–56 (2017).
Kitada, T., DiAndreth, B., Teague, B. & Weiss, R. Programming gene and engineered-cell therapies with synthetic biology. Science 359, eaad1067 (2018).
Neurath, H. & Walsh, K. A. Role of proteolytic enzymes in biological regulation (a review). Proc. Natl Acad. Sci. USA 73, 3825–3832 (1976).
Strasser, A., O’Connor, L. & Dixit, V. M. Apoptosis signaling. Annu. Rev. Biochem. 69, 217–245 (2000).
Kovall, R. A., Gebelein, B., Sprinzak, D. & Kopan, R. The canonical notch signaling pathway: structural and biochemical insights into shape, sugar, and force. Dev. Cell. 41, 228–241 (2017).
Nishimura, K., Fukagawa, T., Takisawa, H., Kakimoto, T. & Kanemaki, M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat. Methods 6, 917–922 (2009).
Fernandez-Rodriguez, J. & Voigt, C. A. Post-translational control of genetic circuits using Potyvirus proteases. Nucleic Acids Res. 44, 6493–6502 (2016).
Majerle, A., Gaber, R., Benčina, M. & Jerala, R. Function-based mutation-resistant synthetic signaling device activated by HIV-1 proteolysis. ACS Synth. Biol. 4, 667–672 (2015).
Schwarz, K. A., Daringer, N. M., Dolberg, T. B. & Leonard, J. N. Rewiring human cellular input–output using modular extracellular sensors. Nat. Chem. Biol. 13, 202–209 (2017).
Shekhawat, S. S., Porter, J. R., Sriprasad, A. & Ghosh, I. An autoinhibited coiled-coil design strategy for split-protein protease sensors. J. Am. Chem. Soc. 131, 15284–15290 (2009).
Stein, V. & Alexandrov, K. Protease-based synthetic sensing and signal amplification. Proc. Natl Acad. Sci. USA 111, 15934–15939 (2014).
Zheng, N. et al. Specific and efficient cleavage of fusion proteins by recombinant plum pox virus NIa protease. Protein Expr. Purif. 57, 153–162 (2008).
Yi, L. et al. Engineering of TEV protease variants by yeast ER sequestration screening (YESS) of combinatorial libraries. Proc. Natl Acad. Sci. USA 110, 7229–7234 (2013).
Seo, J.-K., Choi, H.-S. & Kim, K.-H. Engineering of soybean mosaic virus as a versatile tool for studying protein-protein interactions in soybean. Sci. Rep. 6, 22436 (2016).
Kanno, A., Yamanaka, Y., Hirano, H., Umezawa, Y. & Ozawa, T. Cyclic luciferase for real-time sensing of caspase-3 activities in living mammals. Angew. Chem. Int. Ed. Engl. 46, 7595–7599 (2007).
Nunn, C. M. et al. Crystal structure of the active enzyme and its inactive mutant. Russ. J. Bioorganic Chem. 29, 415–418 (2003).
Wehr, M. C. et al. Monitoring regulated protein-protein interactions using split TEV. Nat. Methods 3, 985–993 (2006).
Liang, F.-S., Ho, W. Q. & Crabtree, G. R. Engineering the ABA plant stress pathway for regulation of induced proximity. Sci. Signal. 4, rs2 (2011).
Gradišar, H. & Jerala, R. De novo design of orthogonal peptide pairs forming parallel coiled-coil heterodimers. J. Pept. Sci. 17, 100–106 (2011).
Woolfson, D. N. The design of coiled-coil structures and assemblies. Adv. Protein Chem. 70, 79–112 (2005).
Grigoryan, G. & Keating, A. E. Structural specificity in coiled-coil interactions. Curr. Opin. Struct. Biol. 18, 477–483 (2008).
Reinke, A. W., Grant, R. A. & Keating, A. E. A synthetic coiled-coil interactome provides heterospecific modules for molecular engineering. J. Am. Chem. Soc. 132, 6025–6031 (2010).
Gradišar, H. et al. Design of a single-chain polypeptide tetrahedron assembled from coiled-coil segments. Nat. Chem. Biol. 9, 362–366 (2013).
Ljubetič, A. et al. Design of coiled-coil protein-origami cages that self-assemble in vitro and in vivo. Nat. Biotechnol. 35, 1094–1101 (2017).
Crick, F. H. C. Is α-keratin a coiled coil? Nature 170, 882–883 (1952).
Lupas, A. Coiled coils: new structures and new functions. Trends. Biochem. Sci. 21, 375–382 (1996).
Berrington, W. R., Smith, K. D., Skerrett, S. J. & Hawn, T. R. Nucleotide-binding oligomerization domain containing-like receptor family, caspase recruitment domain (CARD) containing 4 (NLRC4) regulates intrapulmonary replication of aerosolized Legionella pneumophila. BMC Infect. Dis. 13, 371 (2013).
Oakley, M. G. & Hollenbeck, J. J. The design of antiparallel coiled coils. Curr. Opin. Struct. Biol. 11, 450–457 (2001).
Mittl, P. R. E. et al. The retro-GCN4 leucine zipper sequence forms a stable three-dimensional structure. Proc. Natl Acad. Sci. USA 97, 2562–2566 (2000).
Drobnak, I., Gradišar, H., Ljubetič, A., Merljak, E. & Jerala, R. Modulation of coiled-coil dimer stability through surface residues while preserving pairing specificity. J. Am. Chem. Soc. 139, 8229–8236 (2017).
Pace, C. N. & Scholtz, J. M. A helix propensity scale based on experimental studies of peptides and proteins. Biophys. J. 75, 422–427 (1998).
Phan, J. et al. Structural basis for the substrate specificity of tobacco etch virus protease. J. Biol. Chem. 277, 50564–50572 (2002).
Riechmann, J. L., Laín, S. & García, J. A. Highlights and prospects of potyvirus molecular biology. J. Gen. Virol. 73, 1–16 (1992).
Gao, X. J., Chong, L. S., Kim, M. S. & Elowitz, M. B. Programmable protein circuits in living cells. Science 361, 1252–1258 (2018).
Gibson, D. G. et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009).
Gao, Y. et al. Complex transcriptional modulation with orthogonal and inducible dCas9 regulators. Nat. Methods 13, 1043–1049 (2016).
Iwai, H., Züger, S., Jin, J. & Tam, P.-H. Highly efficient protein trans-splicing by a naturally split DnaE intein from Nostoc punctiforme. FEBS Lett. 580, 1853–1858 (2006).
Lacroix, E., Viguera, A. R. & Serrano, L. Elucidating the folding problem of alpha-helices: local motifs, long-range electrostatics, ionic-strength dependence and prediction of NMR parameters. J. Mol. Biol. 284, 173–191 (1998).
Acknowledgements
The idea and proof of principle for this work were conceived as part of the Slovenian iGEM 2016 project, and members of the team who are not among the authors—M. Meško, M. Mraz, M. Moškon, D. Križaj, R. Krese, N. Franko, L. Magdevska, M. Gradišek, Ž. Pušnik, S. Roškar and K. Cerović—are acknowledged for their contribution. The project was funded by the Slovenian Research Agency (P4-0176 and J3-7034) and an ERC project MaCChines (to R.J.). We thank N. Landau (Division of AIDS, NIAID)) for providing materials.
Author information
Authors and Affiliations
Contributions
T.F., J.L., A.P., T.P., K.L., N.J. and E.M. performed the experiments and analyzed the results; T.F., J.L. and T.L. designed SPOC logic gates; Ž.S. and F.L. designed antiparallel CCs; M.B., T.F., J.L. and R.J. wrote and edited the manuscript; M.B. supervised the experimental work; and R.J. conceptualized the study and acquired funding.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1–8 and Supplementary Figures 1–15
Rights and permissions
About this article
Cite this article
Fink, T., Lonzarić, J., Praznik, A. et al. Design of fast proteolysis-based signaling and logic circuits in mammalian cells. Nat Chem Biol 15, 115–122 (2019). https://doi.org/10.1038/s41589-018-0181-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-018-0181-6
This article is cited by
-
Integrated compact regulators of protein activity enable control of signaling pathways and genome-editing in vivo
Cell Discovery (2024)
-
An orthogonalized PYR1-based CID module with reprogrammable ligand-binding specificity
Nature Chemical Biology (2024)
-
A single-component, light-assisted uncaging switch for endoproteolytic release
Nature Chemical Biology (2024)
-
Irreversible light-activated SpyLigation mediates split-protein assembly in 4D
Nature Protocols (2024)
-
Programmable synthetic receptors: the next-generation of cell and gene therapies
Signal Transduction and Targeted Therapy (2024)