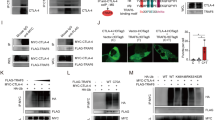
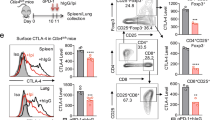
Abstract
Expression of programmed cell death 1 (PD-1) ligand 1 (PD-L1) protects tumor cells from T cell–mediated immune surveillance, and immune checkpoint blockade (ICB) therapies targeting PD-1 and PD-L1 have exhibited significant clinical benefits. However, the relatively low response rate and observed ICB resistance highlight the need to understand the molecular regulation of PD-L1. Here we show that HIP1R targets PD-L1 to lysosomal degradation to alter T cell–mediated cytotoxicity. HIP1R physically interacts with PD-L1 and delivers PD-L1 to the lysosome through a lysosomal targeting signal. Depletion of HIP1R in tumor cells caused PD-L1 accumulation and suppressed T cell–mediated cytotoxicity. A rationally designed peptide (PD-LYSO) incorporating the lysosome-sorting signal and the PD-L1-binding sequence of HIP1R successfully depleted PD-L1 expression in tumor cells. Our results identify the molecular machineries governing the lysosomal degradation of PD-L1 and exemplify the development of a chimeric peptide for targeted degradation of PD-L1 as a crucial anticancer target.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Sonpavde, G. PD-1 and PD-L1 inhibitors as salvage therapy for urothelial carcinoma. N. Engl. J. Med. 376, 1073–1074 (2017).
Chang, Z. L. et al. Rewiring T-cell responses to soluble factors with chimeric antigen receptors. Nat. Chem. Biol. 14, 317–324 (2018).
Brahmer, J. R. et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N. Engl. J. Med. 366, 2455–2465 (2012).
Zerdes, I., Matikas, A., Bergh, J., Rassidakis, G. Z. & Foukakis, T. Genetic, transcriptional and post-translational regulation of the programmed death protein ligand 1 in cancer: biology and clinical correlations. Oncogene 37, 4639–4661 (2018).
Wang, Y. et al. Regulation of PD-L1: emerging routes for targeting tumor immune evasion. Front. Pharmacol. 9, 536 (2018).
Yao, H., Wang, H., Li, C., Fang, J. Y. & Xu, J. Cancer cell-intrinsic PD-1 and implications in combinatorial immunotherapy. Front. Immunol. 9, 1774 (2018).
Santoni, M., Montironi, R. & Battelli, N. Immune checkpoint blockade in advanced renal-cell carcinoma. N. Engl. J. Med. 379, 91–92 (2018).
Yarchoan, M., Hopkins, A. & Jaffee, E. M. Tumor mutational burden and response rate to PD-1 inhibition. N. Engl. J. Med. 377, 2500–2501 (2017).
Maj, T. et al. Oxidative stress controls regulatory T cell apoptosis and suppressor activity and PD-L1-blockade resistance in tumor. Nat. Immunol. 18, 1332–1341 (2017).
Snyder, A. et al. Contribution of systemic and somatic factors to clinical response and resistance to PD-L1 blockade in urothelial cancer: an exploratory multi-omic analysis. PLoS. Med. 14, e1002309 (2017).
Haratake, N. et al. Positive conversion of PD-L1 expression after treatments with chemotherapy and nivolumab. Anticancer Res. 37, 5713–5717 (2017).
Chowdhury, S. et al. Programmed death-ligand 1 overexpression is a prognostic marker for aggressive papillary thyroid cancer and its variants. Oncotarget 7, 32318–32328 (2016).
Burr, M. L. et al. CMTM6 maintains the expression of PD-L1 and regulates anti-tumour immunity. Nature 549, 101–105 (2017).
Dorand, R. D. et al. Cdk5 disruption attenuates tumor PD-L1 expression and promotes antitumor immunity. Science 353, 399–403 (2016).
Kortlever, R. M. et al. Myc cooperates with Ras by programming inflammation and immune suppression. Cell 171, 1301–1315.e14 (2017).
Kataoka, K. et al. Aberrant PD-L1 expression through 3′-UTR disruption in multiple cancers. Nature 534, 402–406 (2016).
Zhang, J. et al. Cyclin D-CDK4 kinase destabilizes PD-L1 via cullin 3-SPOP to control cancer immune surveillance. Nature 553, 91–95 (2018).
Li, C. W. et al. Glycosylation and stabilization of programmed death ligand-1 suppresses T-cell activity. Nat. Commun. 7, 12632 (2016).
Lim, S. O. et al. Deubiquitination and stabilization of PD-L1 by CSN5. Cancer Cell 30, 925–939 (2016).
Chen, G. et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature 560, 382–386 (2018).
Yang, Y. et al. Exosomal PD-L1 harbors active defense function to suppress T cell killing of breast cancer cells and promote tumor growth. Cell Res. 28, 862–864 (2018).
Cha, J. H. et al. Metformin promotes antitumor immunity via endoplasmic-reticulum-associated degradation of PD-L1. Mol. Cell 71, 606–620.e7 (2018).
Mezzadra, R. et al. Identification of CMTM6 and CMTM4 as PD-L1 protein regulators. Nature 549, 106–110 (2017).
Bondeson, D. P. et al. Catalytic in vivo protein knockdown by small-molecule PROTACs. Nat. Chem. Biol. 11, 611–617 (2015).
Sakamoto, K. M. et al. Protacs: chimeric molecules that target proteins to the Skp1-Cullin-F box complex for ubiquitination and degradation. Proc. Natl Acad. Sci. USA 98, 8554–8559 (2001).
Nabet, B. et al. The dTAG system for immediate and target-specific protein degradation. Nat. Chem. Biol. 14, 431–441 (2018).
Bauer, P. O. et al. Harnessing chaperone-mediated autophagy for the selective degradation of mutant huntingtin protein. Nat. Biotechnol. 28, 256–263 (2010).
Fan, X., Jin, W. Y., Lu, J., Wang, J. & Wang, Y. T. Rapid and reversible knockdown of endogenous proteins by peptide-directed lysosomal degradation. Nat. Neurosci. 17, 471–480 (2014).
Bondeson, D. P. et al. Lessons in PROTAC design from selective degradation with a promiscuous warhead. Cell Chem. Biol. 25, 78–87.e5 (2018).
Gadd, M. S. et al. Structural basis of PROTAC cooperative recognition for selective protein degradation. Nat. Chem. Biol. 13, 514–521 (2017).
Van Coillie, S. et al. OncoBinder facilitates interpretation of proteomic interaction data by capturing coactivation pairs in cancer. Oncotarget 7, 17608–17615 (2016).
Deng, R. et al. B7H1/CD80 interaction augments PD-1-dependent T cell apoptosis and ameliorates graft-versus-host disease. J. Immunol. 194, 560–574 (2015).
Wang, X. F. et al. PD-1/PDL1 and CD28/CD80 pathways modulate natural killer T cell function to inhibit hepatitis B virus replication. J. Viral Hepat. 20, 27–39 (2013). Suppl 1.
Gottfried, I., Ehrlich, M. & Ashery, U. The Sla2p/HIP1/HIP1R family: similar structure, similar function in endocytosis? Biochem. Soc. Trans. 38, 187–191 (2010).
Jain, R. N. et al. Hip1r is expressed in gastric parietal cells and is required for tubulovesicle formation and cell survival in mice. J. Clin. Invest. 118, 2459–2470 (2008).
Juneja, V. R. et al. PD-L1 on tumor cells is sufficient for immune evasion in immunogenic tumors and inhibits CD8 T cell cytotoxicity. J. Exp. Med. 214, 895–904 (2017).
Li, C. W. et al. Eradication of triple-negative breast cancer cells by targeting glycosylated PD-L1. Cancer Cell. 33, 187–201.e10 (2018).
Negi, S. et al. LocSigDB: a database of protein localization signals. Database (Oxford) 2015, bav003 (2015).
Greenberg, M., DeTulleo, L., Rapoport, I., Skowronski, J. & Kirchhausen, T. A dileucine motif in HIV-1 Nef is essential for sorting into clathrin-coated pits and for downregulation of CD4. Curr. Biol. 8, 1239–1242 (1998).
Kyttälä, A., Yliannala, K., Schu, P., Jalanko, A. & Luzio, J. P. AP-1 and AP-3 facilitate lysosomal targeting of Batten disease protein CLN3 via its dileucine motif. J. Biol. Chem. 280, 10277–10283 (2005).
Nesbit, M. A. et al. Mutations in AP2S1 cause familial hypocalciuric hypercalcemia type 3. Nat. Genet. 45, 93–97 (2013).
Kantheti, P. et al. Mutation in AP-3 delta in the mocha mouse links endosomal transport to storage deficiency in platelets, melanosomes, and synaptic vesicles. Neuron 21, 111–122 (1998).
Ohno, H. et al. The medium subunits of adaptor complexes recognize distinct but overlapping sets of tyrosine-based sorting signals. J. Biol. Chem. 273, 25915–25921 (1998).
Lundmark, R. & Carlsson, S. R. The beta-appendages of the four adaptor-protein (AP) complexes: structure and binding properties, and identification of sorting nexin 9 as an accessory protein to AP-2. Biochem. J. 362, 597–607 (2002).
Holloway, Z. G. et al. Trafficking of the Menkes copper transporter ATP7A is regulated by clathrin-, AP-2-, AP-1-, and Rab22-dependent steps. Mol. Biol. Cell 24, 1735–1748 S1–8 (2013).
Amorim, N. A. et al. Interaction of HIV-1 Nef protein with the host protein Alix promotes lysosomal targeting of CD4 receptor. J. Biol. Chem. 289, 27744–27756 (2014).
Zhai, Q., Landesman, M. B., Robinson, H., Sundquist, W. I. & Hill, C. P. Identification and structural characterization of the ALIX-binding late domains of simian immunodeficiency virus SIVmac239 and SIVagmTan-1. J. Virol. 85, 632–637 (2011).
Dores, M. R. et al. AP-3 regulates PAR1 ubiquitin-independent MVB/lysosomal sorting via an ALIX-mediated pathway. Mol. Biol. Cell 23, 3612–3623 (2012).
Dores, M. R., Grimsey, N. J., Mendez, F. & Trejo, J. ALIX regulates the ubiquitin-independent lysosomal sorting of the P2Y1 purinergic receptor via a YPX3L motif. PLoS One 11, e0157587 (2016).
Yi, X. et al. Alix (AIP1) is a vasopressin receptor (V2R)-interacting protein that increases lysosomal degradation of the V2R. Am. J. Physiol. Renal Physiol. 292, F1303–F1313 (2007).
Liang, L. et al. A designed peptide targets two types of modifications of p53 with anti-cancer activity. Cell Chem. Biol. 25, 761–774.e765 (2018).
Zhang, Y. et al. Proteomic identification of ERP29 as a key chemoresistant factor activated by the aggregating p53 mutant Arg282Trp. Oncogene 36, 5473–5483 (2017).
Jiao, S. et al. PARP inhibitor upregulates PD-L1 expression and enhances cancer-associated immunosuppression. Clin. Cancer Res. 23, 3711–3720 (2017).
Wang, J. et al. ArhGAP30 promotes p53 acetylation and function in colorectal cancer. Nat. Commun. 5, 4735 (2014).
Acknowledgements
We thank J. Zheng in Shanghai Jiao Tong University and X. Su in Peking University for their inspiring discussions and critical reading of the manuscript. This project was supported by the following grants to J.X.: National Key Research & Development (R&D) Plan (2016YFC0906002); National Natural Science Foundation of China (81874050, 81572326, 81322036, 81320108024); Top-Notch Young Talents Program of China (ZTZ2015-48); Shanghai Municipal Education Commission-Gaofeng Clinical Medicine Grant Support (20152514); “Shu Guang” project supported by Shanghai Municipal Education Commission and Shanghai Education Development Foundation (15SG16); Tang Scholar (SJTU-JX).
Author information
Authors and Affiliations
Contributions
H.W. performed co-immunoprecipitation, cycloheximide-chase, immunofluorescence, flow cytometry, cell proliferation assay, and T cell cytotoxicity assays; H.W., Z.L., H.Y., C.L., H.S., J.L., and J.X. collaboratively performed the other biochemical and cellular experiments including molecular cloning, GST pull-down, peptide binding assay, and PD-1 binding assay; H.W., H.Y., Y.Z., L.L. and J.-Y.F. analyzed flow cytometry data and colocalization between PD-L1 and different subcellular organelles; J.X. reprogramed the OncoBinder package and designed the PD-LYSO peptide. H.W. and J.X. wrote the paper. J.X. conceived and supervised the study.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Table 1, Supplementary Figures 1–11
Rights and permissions
About this article
Cite this article
Wang, H., Yao, H., Li, C. et al. HIP1R targets PD-L1 to lysosomal degradation to alter T cell–mediated cytotoxicity. Nat Chem Biol 15, 42–50 (2019). https://doi.org/10.1038/s41589-018-0161-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-018-0161-x
This article is cited by
-
The application of nanoparticles-based ferroptosis, pyroptosis and autophagy in cancer immunotherapy
Journal of Nanobiotechnology (2024)
-
Pharmaceutical targeting of OTUB2 sensitizes tumors to cytotoxic T cells via degradation of PD-L1
Nature Communications (2024)
-
Carnosine regulation of intracellular pH homeostasis promotes lysosome-dependent tumor immunoevasion
Nature Immunology (2024)
-
Ezurpimtrostat, A Palmitoyl-Protein Thioesterase-1 Inhibitor, Combined with PD-1 Inhibition Provides CD8+ Lymphocyte Repopulation in Hepatocellular Carcinoma
Targeted Oncology (2024)
-
Comprehensive assessment of TECENTRIQ® and OPDIVO®: analyzing immunotherapy indications withdrawn in triple-negative breast cancer and hepatocellular carcinoma
Cancer and Metastasis Reviews (2024)