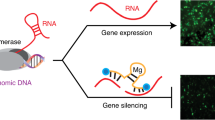
Abstract
Synthetic mRNA is an attractive vehicle for gene therapies because of its transient nature and improved safety profile over DNA. However, unlike DNA, broadly applicable methods to control expression from mRNA are lacking. Here we describe a platform for small-molecule-based regulation of expression from modified RNA (modRNA) and self-replicating RNA (replicon) delivered to mammalian cells. Specifically, we engineer small-molecule-responsive RNA binding proteins to control expression of proteins from RNA-encoded genetic circuits. Coupled with specific modRNA dosages or engineered elements from a replicon, including a subgenomic promoter library, we demonstrate the capability to externally regulate the timing and level of protein expression. These control mechanisms facilitate the construction of ON, OFF, and two-output switches, with potential therapeutic applications such as inducible cancer immunotherapies. These circuits, along with other synthetic networks that can be developed using these tools, will expand the utility of synthetic mRNA as a therapeutic modality.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The authors declare that data supporting the finding of this study are available within the article and its Supplementary Information. Sample analysis of cytometry data can be found in Supplementary Fig. 29. Replicon MoClo assembly plasmids have been submitted to Addgene with the accession numbers 115928, 115929, 115930, 115931, 115932, 115933, 115934, 115935, 115936, 115937, 115938, 115939, 115940, 115941, 115942, 115943, 115944, 115945, 115946, 115947, 115948, 115949, 115950, 115951, 115952, 115953, 115954, 115955, 115956, 115957, 115958, 115959, 115960, 115961, 115962, 115963, 115964, 115965, 115966, and 115967. A more detailed table can be found in Supplementary Fig. 30. Additional data are available from the corresponding authors upon reasonable request.
References
Naldini, L. Gene therapy returns to centre stage. Nature 526, 351–360 (2015).
Rangarajan, S. et al. AAV5-factor VIII gene transfer in severe hemophilia A. N. Engl. J. Med. 377, 2519–2530 (2017).
George, L. A. et al. Hemophilia B gene therapy with a high-specific-activity factor IX variant. N. Engl. J. Med. 377, 2215–2227 (2017).
Sahin, U., Karikó, K. & Türeci, Ö. mRNA-based therapeutics–developing a new class of drugs. Nat. Rev. Drug Discov. 13, 759–780 (2014).
Karikó, K., Buckstein, M., Ni, H. & Weissman, D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity 23, 165–175 (2005).
Karikó, K. et al. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther. 16, 1833–1840 (2008).
Ljungberg, K. & Liljeström, P. Self-replicating alphavirus RNA vaccines. Expert Rev. Vaccines 14, 177–194 (2015).
Strauss, J. H. & Strauss, E. G. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 58, 491–562 (1994).
Kitada, T., DiAndreth, B., Teague, B. & Weiss, R. Programming gene and engineered-cell therapies with synthetic biology. Science 359, eaad1067 (2018).
Jenkins, P. V., Rawley, O., Smith, O. P. & O’Donnell, J. S. Elevated factor VIII levels and risk of venous thrombosis. Br. J. Haematol. 157, 653–663 (2012).
Heikal, N. M. et al. Elevated factor IX activity is associated with an increased odds ratio for both arterial and venous thrombotic events. Am. J. Clin. Pathol. 140, 680–685 (2013).
Leonard, J. P. et al. Effects of single-dose interleukin-12 exposure on interleukin-12-associated toxicity and interferon-gamma production. Blood 90, 2541–2548 (1997).
Ryding, A. D., Sharp, M. G. & Mullins, J. J. Conditional transgenic technologies. J. Endocrinol. 171, 1–14 (2001).
Townshend, B., Kennedy, A. B., Xiang, J. S. & Smolke, C. D. High-throughput cellular RNA device engineering. Nat. Methods 12, 989–994 (2015).
Banaszynski, L. A., Chen, L. C., Maynard-Smith, L. A., Ooi, A. G. L. & Wandless, T. J. A. A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules. Cell 126, 995–1004 (2006).
Bonger, K. M., Chen, L. C., Liu, C. W. & Wandless, T. J. Small-molecule displacement of a cryptic degron causes conditional protein degradation. Nat. Chem. Biol. 7, 531–537 (2011).
Chung, H. K. et al. Tunable and reversible drug control of protein production via a self-excising degron. Nat. Chem. Biol. 11, 713–720 (2015).
An, W. et al. Engineering FKBP-based destabilizing domains to build sophisticated protein regulation systems. PLoS One 10, e0145783 (2015).
Wroblewska, L. et al. Mammalian synthetic circuits with RNA binding proteins for RNA-only delivery. Nat. Biotechnol. 33, 839–841 (2015).
Kaczmarek, J. C., Kowalski, P. S. & Anderson, D. G. Advances in the delivery of RNA therapeutics: from concept to clinical reality. Genome Med. 9, 60 (2017).
Garber, K. Alnylam terminates revusiran program, stock plunges. Nat. Biotechnol. 34, 1213–1214 (2016).
Andries, O. et al. N1-methylpseudouridine-incorporated mRNA outperforms pseudouridine-incorporated mRNA by providing enhanced protein expression and reduced immunogenicity in mammalian cell lines and mice. J. Control. Release 217, 337–344 (2015).
Iwamoto, M., Björklund, T., Lundberg, C., Kirik, D. & Wandless, T. J. A general chemical method to regulate protein stability in the mammalian central nervous system. Chem. Biol. 17, 981–988 (2010).
Sellmyer, M. A., Chen, L. C., Egeler, E. L., Rakhit, R. & Wandless, T. J. Intracellular context affects levels of a chemically dependent destabilizing domain. PLoS One 7, e43297 (2012).
Saito, H. et al. Synthetic translational regulation by an L7Ae-kink-turn RNP switch. Nat. Chem. Biol. 6, 71–78 (2010).
Gossen, M. & Bujard, H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. USA 89, 5547–5551 (1992).
Belmont, B. J. & Niles, J. C. Engineering a direct and inducible protein-RNA interaction to regulate RNA biology. ACS Chem. Biol. 5, 851–861 (2010).
Ganesan, S. M., Falla, A., Goldfless, S. J., Nasamu, A. S. & Niles, J. C. Synthetic RNA-protein modules integrated with native translation mechanisms to control gene expression in malaria parasites. Nat. Commun. 7, 10727 (2016).
Yoshioka, N. et al. Efficient generation of human iPSCs by a synthetic self-replicative RNA. Cell Stem Cell 13, 246–254 (2013).
Lundstrom, K. Alphavirus-based vaccines. Viruses 6, 2392–2415 (2014).
Beal, J. et al. Model-driven engineering of gene expression from RNA replicons. ACS Synth. Biol. 4, 48–56 (2015).
Frolov, I. et al. Alphavirus-based expression vectors: strategies and applications. Proc. Natl. Acad. Sci. USA 93, 11371–11377 (1996).
Levis, R., Schlesinger, S. & Huang, H. V. Promoter for Sindbis virus RNA-dependent subgenomic RNA transcription. J. Virol. 64, 1726–1733 (1990).
Weber, E., Emgler, C., Gruetzner, R., Werner, S. & Marillonnet, S. A modular cloning system for standardized assembly of multigene constructs. PLoS One 6, e16765–1733 (2018).
Culver, J. N., Lehto, K., Close, S. M., Hilf, M. E. & Dawson, W. O. Genomic position affects the expression of tobacco mosaic virus movement and coat protein genes. Proc. Natl. Acad. Sci. USA 90, 2055–2059 (1993).
Andries, O., Kitada, T., Bodner, K., Sanders, N. N. & Weiss, R. Synthetic biology devices and circuits for RNA-based ‘smart vaccines’: a propositional review. Expert Rev. Vaccines 14, 313–331 (2015).
Hoarau, J. J. et al. Identical strength of the T cell responses against E2, nsP1 and capsid CHIKV proteins in recovered and chronic patients after the epidemics of 2005-2006 in La Reunion Island. PLoS One 8, e84695 (2013).
Nathwani, A. C. et al. Long-term safety and efficacy of factor IX gene therapy in hemophilia B. N. Engl. J. Med. 371, 1994–2004 (2014).
Maldonado, R. A. et al. Polymeric synthetic nanoparticles for the induction of antigen-specific immunological tolerance. Proc. Natl. Acad. Sci. USA 112, E156–E165 (2015).
Kishimoto, T. K. et al. Improving the efficacy and safety of biologic drugs with tolerogenic nanoparticles. Nat. Nanotechnol. 11, 890–899 (2016).
Haurwitz, R. E., Jinek, M., Wiedenheft, B., Zhou, K. & Doudna, J. A. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science 329, 1355–1358 (2010).
Engler, C., Gruetzner, R., Kandzia, R. & Marillonnet, S. Golden gate shuffling: a one-pot DNA shuffling method based on type IIs restriction enzymes. PLoS One 4, e5553 (2009).
Wielgosz, M. M., Raju, R. & Huang, H. V. Sequence requirements for Sindbis virus subgenomic mRNA promoter function in cultured cells. J. Virol. 75, 3509–3519 (2001).
Acknowledgements
The authors would like to thank J. Niles, B.J. Belmont, and S. Maddur Ganesan (MIT) for discussions regarding the TetR–aptamer system, A. Ghodasara (MIT) for discussions and technical assistance related to aptamers/aptazymes, and R. Petersen (Designs By Robin) for consultations with figure design and layout. This work was supported by research grants from the Defense Advanced Research Projects Agency (W911NF-11-2-0054: awarded to J.B., D.D., and R.W., supported T.W., J.B.R., X.Z., E.P., J.B., D.D., T.K., and R.W. and W32P4Q-13-1-0011: awarded to R.W., supported X.Z., T.K., and R.W.), the National Science Foundation (NSF#1522074, NSF#1521759: awarded to D.D., supported T.W. and D.D., CCF-1521925: awarded to D.D. and R.W., supported T.W, J.R.B., D.D., and R.W., CNS-1446607: awarded to R.W., supported J.R.B., B.T., and R.W., and MCB-1745645: awarded to R.W., supported J.R.B. and R.W.), the National Institutes of Health (5-R01-CA206218: awarded to R.W., supported J.B.R., E.P., and R.W.), the Ragon Institute of MGH, MIT and Harvard (awarded to R.W., supported A.W. and R.W.), the Special Research Fund from Ghent University (awarded to N.N.S.), and The Research Foundation - Flanders (FWO; G.0235.11N and G.0621.10N: awarded to N.N.S.). This work was also supported by a sponsored research agreement with Crucell Holland B.V. (awarded to R.W., supported: B.D., E.P., and R.W.). We further acknowledge the following support: MIT-Amgen UROP Scholars Program (K.B.), Gabilan Stanford Graduate Fellowship (K.B.), Fannie and John Hertz Foundation Fellowship - Hertz-Draper Fellow (K.B.), Stanford EDGE-STEM Doctoral Fellowship (K.B.), PhD fellowship and international mobility grant from FWO (O.A.), and the Emmanuel van der Schueren fellowship from “Kom op tegen Kanker” (O.A.).
Author information
Authors and Affiliations
Contributions
J.R.B., T.W., K.B., and T.K. designed and performed experiments and analyzed data. X.Z., A.W., E.P., and B.D. performed experiments and interpreted data. O.A., N.N.S., and D.D. designed experiments. B.A. performed experiments. J.B. analyzed data. T.K. and R.W. supervised the study. T.W., J.R.B., B.T., T.K., and R.W. wrote the manuscript with the support of all other authors.
Corresponding authors
Ethics declarations
Competing interests
MIT has filed a patent application (No. 15/509,258) pertaining to the technology described in this paper. T.W., J.R.B., K.B., T.K., and R.W. are co-inventors on this patent application. J.R.B. and T.K. are shareholders of a company founded based on the technology described in this article.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figures 1–30
Rights and permissions
About this article
Cite this article
Wagner, T.E., Becraft, J.R., Bodner, K. et al. Small-molecule-based regulation of RNA-delivered circuits in mammalian cells. Nat Chem Biol 14, 1043–1050 (2018). https://doi.org/10.1038/s41589-018-0146-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-018-0146-9
This article is cited by
-
Customizing cellular signal processing by synthetic multi-level regulatory circuits
Nature Communications (2023)
-
Programmable mammalian translational modulators by CRISPR-associated proteins
Nature Communications (2023)
-
Synthetic neuromorphic computing in living cells
Nature Communications (2022)
-
Caliciviral protein-based artificial translational activator for mammalian gene circuits with RNA-only delivery
Nature Communications (2020)
-
Synthetic Biology Goes Cell-Free
BMC Biology (2019)