Abstract
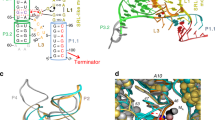
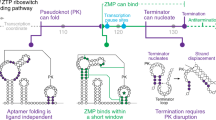
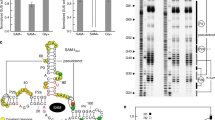
The ykkC family of bacterial riboswitches combines several widespread classes that have similar secondary structures and consensus motifs but control different genes in response to different cellular metabolites. Here we report the crystal structures of two distinct ykkC riboswitches specifically bound to their cognate ligand ppGpp, a second messenger involved in stress response, or PRPP, a precursor in purine biosynthesis. Both RNAs adopt similar structures and contain a conserved core previously observed in the guanidine-specific ykkC riboswitch. However, ppGpp and PRPP riboswitches uniquely employ an additional helical element that joins the ends of the ligand-sensing domains and creates a tunnel for direct and Mg2+-mediated binding of ligands. Mutational and footprinting experiments highlight the importance of conserved nucleotides forming the tunnel and long-distance contacts for ligand binding and genetic response. Our work provides new insights into the specificity of riboswitches and gives a unique opportunity for future studies of RNA evolution.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Serganov, A. & Nudler, E. A decade of riboswitches. Cell 152, 17–24 (2013).
Breaker, R. R. Prospects for riboswitch discovery and analysis. Mol. Cell 43, 867–879 (2011).
Greenlee, E. B. et al. Challenges of ligand identification for the second wave of orphan riboswitch candidates. RNA Biol. 15, 377–390 (2018).
McCown, P. J., Corbino, K. A., Stav, S., Sherlock, M. E. & Breaker, R. R. Riboswitch diversity and distribution. RNA 23, 995–1011 (2017).
Barrick, J. E. et al. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc. Natl Acad. Sci. USA 101, 6421–6426 (2004).
Sherlock, M. E., Sudarsan, N., Stav, S. & Breaker, R. R. Tandem riboswitches form a natural Boolean logic gate to control purine metabolism in bacteria. eLife 7, e33908 (2018).
Sherlock, M. E., Sudarsan, N. & Breaker, R. R. Riboswitches for the alarmone ppGpp expand the collection of RNA-based signaling systems. Proc. Natl Acad. Sci. USA 115, 6052–6057 (2018).
Nelson, J. W., Atilho, R. M., Sherlock, M. E., Stockbridge, R. B. & Breaker, R. R. Metabolism of free guanidine in bacteria is regulated by a widespread riboswitch class. Mol. Cell 65, 220–230 (2017).
Hove-Jensen, B. et al. Phosphoribosyl diphosphate (PRPP): biosynthesis, enzymology, utilization, and metabolic significance. Microbiol. Mol. Biol. Rev. 81, e00040–e16 (2016).
Sudarsan, N. et al. Tandem riboswitch architectures exhibit complex gene control functions. Science 314, 300–304 (2006).
Hauryliuk, V., Atkinson, G. C., Murakami, K. S., Tenson, T. & Gerdes, K. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat. Rev. Microbiol. 13, 298–309 (2015).
Steinchen, W. & Bange, G. The magic dance of the alarmones (p)ppGpp. Mol. Microbiol. 101, 531–544 (2016).
Reiss, C. W., Xiong, Y. & Strobel, S. A. Structural basis for ligand binding to the guanidine-I riboswitch. Structure 25, 195–202 (2017).
Battaglia, R. A., Price, I. R. & Ke, A. Structural basis for guanidine sensing by the ykkC family of riboswitches. RNA 23, 578–585 (2017).
Adams, P. D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr 58, 1948–1954 (2002).
Sokoloski, J. E., Godfrey, S. A., Dombrowski, S. E. & Bevilacqua, P. C. Prevalence of syn nucleobases in the active sites of functional RNAs. RNA 17, 1775–1787 (2011).
Serganov, A., Huang, L. & Patel, D. J. Structural insights into amino acid binding and gene control by a lysine riboswitch. Nature 455, 1263–1267 (2008).
Huang, L., Serganov, A. & Patel, D. J. Structural insights into ligand recognition by a sensing domain of the cooperative glycine riboswitch. Mol. Cell 40, 774–786 (2010).
Garst, A. D., Héroux, A., Rambo, R. P. & Batey, R. T. Crystal structure of the lysine riboswitch regulatory mRNA element. J. Biol. Chem. 283, 22347–22351 (2008).
Vicens, Q., Mondragón, E. & Batey, R. T. Molecular sensing by the aptamer domain of the FMN riboswitch: a general model for ligand binding by conformational selection. Nucleic Acids Res. 39, 8586–8598 (2011).
Serganov, A., Polonskaia, A., Phan, A. T., Breaker, R. R. & Patel, D. J. Structural basis for gene regulation by a thiamine pyrophosphate-sensing riboswitch. Nature 441, 1167–1171 (2006).
Serganov, A., Huang, L. & Patel, D. J. Coenzyme recognition and gene regulation by a flavin mononucleotide riboswitch. Nature 458, 233–237 (2009).
Thore, S., Leibundgut, M. & Ban, N. Structure of the eukaryotic thiamine pyrophosphate riboswitch with its regulatory ligand. Science 312, 1208–1211 (2006).
Edwards, T. E. & Ferré-D’Amaré, A. R. Crystal structures of the thi-box riboswitch bound to thiamine pyrophosphate analogs reveal adaptive RNA-small molecule recognition. Structure 14, 1459–1468 (2006).
Klein, D. J. & Ferré-D’Amaré, A. R. Structural basis of glmS ribozyme activation by glucosamine-6-phosphate. Science 313, 1752–1756 (2006).
Cochrane, J. C., Lipchock, S. V. & Strobel, S. A. Structural investigation of the GlmS ribozyme bound to Its catalytic cofactor. Chem. Biol. 14, 97–105 (2007).
Ren, A. & Patel, D. J. c-di-AMP binds the ydaO riboswitch in two pseudo-symmetry-related pockets. Nat. Chem. Biol. 10, 780–786 (2014).
Mandal, M. & Breaker, R. R. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nat. Struct. Mol. Biol. 11, 29–35 (2004).
Serganov, A. et al. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chem. Biol. 11, 1729–1741 (2004).
Mandal, M., Boese, B., Barrick, J. E., Winkler, W. C. & Breaker, R. R. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell 113, 577–586 (2003).
Edwards, A. L. & Batey, R. T. A structural basis for the recognition of 2′-deoxyguanosine by the purine riboswitch. J. Mol. Biol. 385, 938–948 (2009).
Pikovskaya, O., Polonskaia, A., Patel, D. J. & Serganov, A. Structural principles of nucleoside selectivity in a 2′-deoxyguanosine riboswitch. Nat. Chem. Biol. 7, 748–755 (2011).
Kim, J. N., Roth, A. & Breaker, R. R. Guanine riboswitch variants from Mesoplasma florum selectively recognize 2'-deoxyguanosine. Proc. Natl Acad. Sci. USA 104, 16092–16097 (2007).
Regulski, E. E. et al. A widespread riboswitch candidate that controls bacterial genes involved in molybdenum cofactor and tungsten cofactor metabolism. Mol. Microbiol. 68, 918–932 (2008).
Nelson, J. W. et al. Control of bacterial exoelectrogenesis by c-AMP-GMP. Proc. Natl Acad. Sci. USA 112, 5389–5394 (2015).
Ren, A. et al. Structural basis for molecular discrimination by a 3′,3′-cGAMP sensing riboswitch. Cell Reports 11, 1–12 (2015).
Smith, K. D., Lipchock, S. V., Livingston, A. L., Shanahan, C. A. & Strobel, S. A. Structural and biochemical determinants of ligand binding by the c-di-GMP riboswitch. Biochemistry 49, 7351–7359 (2010).
Kellenberger, C. A. et al. GEMM-I riboswitches from Geobacter sense the bacterial second messenger cyclic AMP-GMP. Proc. Natl Acad. Sci. USA 112, 5383–5388 (2015).
Liu, K., Bittner, A. N. & Wang, J. D. Diversity in (p)ppGpp metabolism and effectors. Curr. Opin. Microbiol. 24, 72–79 (2015).
Reiss, C. W. & Strobel, S. A. Structural basis for ligand binding to the guanidine-II riboswitch. RNA 23, 1338–1343 (2017).
Huang, L., Wang, J. & Lilley, D. M. J. The structure of the guanidine-II riboswitch. Cell Chem. Biol. 24, 695–702.e2 (2017).
Huang, L., Wang, J., Wilson, T. J. & Lilley, D. M. J. Structure of the guanidine III riboswitch. Cell Chem. Biol. 24, 1407–1415.e2 (2017).
Yao, Z. et al. A computational pipeline for high- throughput discovery of cis-regulatory noncoding RNA in prokaryotes. PLOS Comput. Biol. 3, e126 (2007).
Nawrocki, E. P. & Eddy, S. R. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 29, 2933–2935 (2013).
Barrick, J. E. Predicting riboswitch regulation on a genomic scale. Methods Mol. Biol. 540, 1–13 (2009).
Peselis, A., Gao, A. & Serganov, A. Preparation and crystallization of riboswitches. Methods Mol. Biol. 1320, 21–36 (2016).
Karplus, P. A. & Diederichs, K. Assessing and maximizing data quality in macromolecular crystallography. Curr. Opin. Struct. Biol. 34, 60–68 (2015).
Terwilliger, T. C. et al. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D Biol. Crystallogr. 64, 61–69 (2008).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Afonine, P. V. et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 (2012).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 (1997).
Acknowledgements
We thank D. Fenyo for help with bioinformatics searches. We thank personnel of beamlines 17-ID-2 at the Brookhaven National Laboratory and 24-ID at the Argonne National Laboratory for help with data collection. This research was supported by the NIH grants R01GM112940 to A.S., F31GM119357 and T32 GM88118 to A.P. This work uses the Northeastern Collaborative Access Team beamlines, which are funded by the National Institutes of Health (NIH) grants (P41 GM103403 and S10 RR029205) and resources of the Advanced Photon Source, operated for the DOE Office of Science under Contract no. DE-AC02-06CH11357. The beamline 17-ID-2 is supported in part by the DOE Office of Biological and Environmental Research (KP1605010, KC0401040) and by the NIH (P41GM111244).
Author information
Authors and Affiliations
Contributions
A.P. crystallized the riboswitches, determined their structures and conducted biochemical experiments. A.S. contributed to determination and refinement of the structures. A.P. and A.S. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1 and 2, Supplementary Figures 1–8
Rights and permissions
About this article
Cite this article
Peselis, A., Serganov, A. ykkC riboswitches employ an add-on helix to adjust specificity for polyanionic ligands. Nat Chem Biol 14, 887–894 (2018). https://doi.org/10.1038/s41589-018-0114-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-018-0114-4
This article is cited by
-
Observation of coordinated RNA folding events by systematic cotranscriptional RNA structure probing
Nature Communications (2023)
-
Structure and mechanism of a methyltransferase ribozyme
Nature Chemical Biology (2022)
-
Structural basis of amino acid surveillance by higher-order tRNA-mRNA interactions
Nature Structural & Molecular Biology (2019)