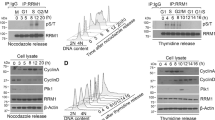
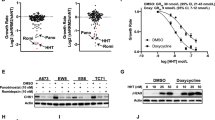
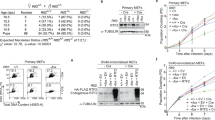
Abstract
Since the origins of DNA-based life, the enzyme ribonucleotide reductase (RNR) has spurred proliferation because of its rate-limiting role in de novo deoxynucleoside-triphosphate (dNTP) biosynthesis. Paradoxically, the large subunit, RNR-α, of this obligatory two-component complex in mammals plays a context-specific antiproliferative role. There is little explanation for this dichotomy. Here, we show that RNR-α has a previously unrecognized DNA-replication inhibition function, leading to growth retardation. This underappreciated biological activity functions in the nucleus, where RNR-α interacts with ZRANB3. This process suppresses ZRANB3’s function in unstressed cells, which we show to promote DNA synthesis. This nonreductase function of RNR-α is promoted by RNR-α hexamerization—induced by a natural and synthetic nucleotide of dA/ClF/CLA/FLU—which elicits rapid RNR-α nuclear import. The newly discovered nuclear signaling axis is a primary defense against elevated or imbalanced dNTP pools that can exert mutagenic effects irrespective of the cell cycle.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
O’Donnell, M., Langston, L. & Stillman, B. Principles and concepts of DNA replication in bacteria, archaea, and eukarya. Cold Spring Harb. Perspect. Biol. 5, a010108 (2013).
Nordlund, P. & Reichard, P. Ribonucleotide reductases. Annu. Rev. Biochem. 75, 681–706 (2006).
Aye, Y., Li, M., Long, M. J. & Weiss, R. S. Ribonucleotide reductase and cancer: biological mechanisms and targeted therapies. Oncogene 34, 2011–2021 (2015).
Aye, Y. & Stubbe, J. Clofarabine 5′-di and -triphosphates inhibit human ribonucleotide reductase by altering the quaternary structure of its large subunit. Proc. Natl. Acad. Sci. USA 108, 9815–9820 (2011).
Fairman, J. W. et al. Structural basis for allosteric regulation of human ribonucleotide reductase by nucleotide-induced oligomerization. Nat. Struct. Mol. Biol. 18, 316–322 (2011).
Cooperman, B. S. & Kashlan, O. B. A comprehensive model for the allosteric regulation of Class Ia ribonucleotide reductases. Adv. Enzyme Regul. 43, 167–182 (2003).
Hofer, A., Crona, M., Logan, D. T. & Sjöberg, B. M. DNA building blocks: keeping control of manufacture. Crit. Rev. Biochem. Mol. Biol. 47, 50–63 (2012).
Aye, Y. et al. Clofarabine targets the large subunit (α) of human ribonucleotide reductase in live cells by assembly into persistent hexamers. Chem. Biol. 19, 799–805 (2012).
Fu, Y., Lin, H., Wisitpitthaya, S., Blessing, W. A. & Aye, Y. A fluorimetric readout reporting the kinetics of nucleotide-induced human ribonucleotide reductase oligomerization. ChemBioChem 15, 2598–2604 (2014).
Wisitpitthaya, S. et al. Cladribine and fludarabine nucleotides induce distinct hexamers defining a common mode of reversible rnr inhibition. ACS Chem. Biol. 11, 2021–2032 (2016).
Ando, N. et al. Allosteric inhibition of human ribonucleotide reductase by datp entails the stabilization of a hexamer. Biochemistry 55, 373–381 (2016).
Brignole, E. J. et al. 3.3-Å resolution cryo-EM structure of human ribonucleotide reductase with substrate and allosteric regulators bound. eLife 7, e31502 (2018).
Rofougaran, R., Vodnala, M. & Hofer, A. Enzymatically active mammalian ribonucleotide reductase exists primarily as an alpha6beta2 octamer. J. Biol. Chem. 281, 27705–27711 (2006).
Engström, Y. & Rozell, B. Immunocytochemical evidence for the cytoplasmic localization and differential expression during the cell cycle of the M1 and M2 subunits of mammalian ribonucleotide reductase. EMBO J. 7, 1615–1620 (1988).
Pontarin, G. et al. Ribonucleotide reduction is a cytosolic process in mammalian cells independently of DNA damage. Proc. Natl. Acad. Sci. USA 105, 17801–17806 (2008).
Fong, K. M., Zimmerman, P. V. & Smith, P. J. Correlation of loss of heterozygosity at 11p with tumour progression and survival in non-small cell lung cancer. Genes Chromosom. Cancer 10, 183–189 (1994).
Wang, Q. et al. Ribonucleotide reductase large subunit M1 predicts poor survival due to modulation of proliferative and invasive ability of gastric cancer. PLoS One 8, e70191 (2013).
Lee, J. J. et al. The immunohistochemical overexpression of ribonucleotide reductase regulatory subunit M1 (RRM1) protein is a predictor of shorter survival to gemcitabine-based chemotherapy in advanced non-small cell lung cancer (NSCLC). Lung Cancer 70, 205–210 (2010).
Mathews, C. K. Deoxyribonucleotide metabolism, mutagenesis and cancer. Nat. Rev. Cancer 15, 528–539 (2015).
Ali, I. U., Lidereau, R., Theillet, C. & Callahan, R. Reduction to homozygosity of genes on chromosome 11 in human breast neoplasia. Science 238, 185–188 (1987).
Cao, M. Y. et al. Adenovirus-mediated ribonucleotide reductase R1 gene therapy of human colon adenocarcinoma. Clin. Cancer Res. 9, 4553–4561 (2003).
Fan, H., Huang, A., Villegas, C. & Wright, J. A. The R1 component of mammalian ribonucleotide reductase has malignancy-suppressing activity as demonstrated by gene transfer experiments. Proc. Natl. Acad. Sci. USA 94, 13181–13186 (1997).
Bepler, G. et al. RRM1 and PTEN as prognostic parameters for overall and disease-free survival in patients with non-small-cell lung cancer. J. Clin. Oncol. 22, 1878–1885 (2004).
Gautam, A. & Bepler, G. Suppression of lung tumor formation by the regulatory subunit of ribonucleotide reductase. Cancer Res. 66, 6497–6502 (2006).
Gautam, A., Li, Z. R. & Bepler, G. RRM1-induced metastasis suppression through PTEN-regulated pathways. Oncogene 22, 2135–2142 (2003).
Pitterle, D. M. et al. Lung cancer and the human gene for ribonucleotide reductase subunit M1 (RRM1). Mamm. Genome 10, 916–922 (1999).
Zheng, Z. et al. DNA synthesis and repair genes RRM1 and ERCC1 in lung cancer. N. Engl. J. Med. 356, 800–808 (2007).
Qi, H. et al. Non-enzymatic action of RRM1 protein upregulates PTEN leading to inhibition of colorectal cancer metastasis. Tumour Biol. 36, 4833–4842 (2015).
Davoli, T. et al. Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell 155, 948–962 (2013).
Ciccia, A. et al. Polyubiquitinated PCNA recruits the ZRANB3 translocase to maintain genomic integrity after replication stress. Mol. Cell 47, 396–409 (2012).
Weston, R., Peeters, H. & Ahel, D. ZRANB3 is a structure-specific ATP-dependent endonuclease involved in replication stress response. Genes Dev. 26, 1558–1572 (2012).
Yuan, J., Ghosal, G. & Chen, J. The HARP-like domain-containing protein AH2/ZRANB3 binds to PCNA and participates in cellular response to replication stress. Mol. Cell 47, 410–421 (2012).
Stubbe, J. & van Der Donk, W. A. Protein radicals in enzyme catalysis. Chem. Rev. 98, 705–762 (1998).
Andersen, P. L., Xu, F. & Xiao, W. Eukaryotic DNA damage tolerance and translesion synthesis through covalent modifications of PCNA. Cell Res. 18, 162–173 (2008).
Sirbu, B. M., Couch, F. B. & Cortez, D. Monitoring the spatiotemporal dynamics of proteins at replication forks and in assembled chromatin using isolation of proteins on nascent DNA. Nat. Protoc. 7, 594–605 (2012).
Gratzner, H. G. Monoclonal antibody to 5-bromo- and 5-iododeoxyuridine: A new reagent for detection of DNA replication. Science 218, 474–475 (1982).
Gilljam, K. M. et al. Identification of a novel, widespread, and functionally important PCNA-binding motif. J. Cell Biol. 186, 645–654 (2009).
Su Lim, C. et al. Measurement of the nucleus area and nucleus/cytoplasm and mitochondria/nucleus ratios in human colon tissues by dual-colour two-photon microscopy imaging. Sci. Rep. 5, 18521 (2015).
Specks, J., Lecona, E., Lopez-Contreras, A. J. & Fernandez-Capetillo, O. A single conserved residue mediates binding of the ribonucleotide reductase catalytic subunit rrm1 to rrm2 and is essential for mouse development. Mol. Cell. Biol. 35, 2910–2917 (2015).
Aye, Y., Long, M. J. & Stubbe, J. Mechanistic studies of semicarbazone triapine targeting human ribonucleotide reductase in vitro and in mammalian cells: tyrosyl radical quenching not involving reactive oxygen species. J. Biol. Chem. 287, 35768–35778 (2012).
Ewald, B., Sampath, D. & Plunkett, W. Nucleoside analogs: molecular mechanisms signaling cell death. Oncogene 27, 6522–6537 (2008).
Niida, H. et al. Essential role of Tip60-dependent recruitment of ribonucleotide reductase at DNA damage sites in DNA repair during G1 phase. Genes Dev. 24, 333–338 (2010).
Tsao, N., Yang, Y. C., Deng, Y. J. & Chang, Z. F. The direct interaction of NME3 with Tip60 in DNA repair. Biochem. J. 473, 1237–1245 (2016).
Bianchi, V., Pontis, E. & Reichard, P. Dynamics of the dATP pool in cultured mammalian cells. Exp. Cell Res. 199, 120–128 (1992).
Arnaoutov, A. & Dasso, M. Enzyme regulation. IRBIT is a novel regulator of ribonucleotide reductase in higher eukaryotes. Science 345, 1512–1515 (2014).
Vujanovic, M. et al. Replication fork slowing and reversal upon DNA damage require pcna polyubiquitination and zranb3 DNA translocase activity. Mol. Cell 67, 882–890.e5 (2017).
Fu, Y. et al. Uncoupling of allosteric and oligomeric regulation in a functional hybrid enzyme constructed from Escherichia coli and human ribonucleotide reductase. Biochemistry 52, 7050–7059 (2013).
Kumar, D., Viberg, J., Nilsson, A. K. & Chabes, A. Highly mutagenic and severely imbalanced dNTP pools can escape detection by the S-phase checkpoint. Nucleic Acids Res. 38, 3975–3983 (2010).
Pai, C. C. & Kearsey, S. E. A critical balance: Dntps and the maintenance of genome stability. Genes (Basel) 8, 57 (2017).
Anglana, M., Apiou, F., Bensimon, A. & Debatisse, M. Dynamics of DNA replication in mammalian somatic cells: nucleotide pool modulates origin choice and interorigin spacing. Cell 114, 385–394 (2003).
Kocsis, E., Cerritelli, M. E., Trus, B. L., Cheng, N. & Steven, A. C. Improved methods for determination of rotational symmetries in macromolecules. Ultramicroscopy 60, 219–228 (1995).
Marabini, R. et al. Xmipp: An image processing package for electron microscopy. J. Struct. Biol. 116, 237–240 (1996).
Scheres, S. H. et al. Maximum-likelihood multi-reference refinement for electron microscopy images. J. Mol. Biol. 348, 139–149 (2005).
Sorzano, C. O. et al. XMIPP: a new generation of an open-source image processing package for electron microscopy. J. Struct. Biol. 148, 194–204 (2004).
Jackson, D. A. & Pombo, A. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J. Cell Biol. 140, 1285–1295 (1998).
Seluanov, A., Mittelman, D., Pereira-Smith, O. M., Wilson, J. H. & Gorbunova, V. DNA end joining becomes less efficient and more error-prone during cellular senescence. Proc. Natl. Acad. Sci. USA 101, 7624–7629 (2004).
Ferraro, P. et al. Mitochondrial deoxynucleotide pools in quiescent fibroblasts: a possible model for mitochondrial neurogastrointestinal encephalomyopathy (MNGIE). J. Biol. Chem. 280, 24472–24480 (2005).
Acknowledgements
Thanks to members of the individual labs who generously provided plasmids and shRNAs as indicated in on-line methods; J. Page for contributing to the creation of RNR-α(D57N) knock-in mice; A. Arnaoutov (Eunice Kennedy Shriver National Institute of Child Health and Human Development, NIH) for pfastbac-GST-IRBIT; V. Gorbunova (University of Rochester) for NHEJ-reporter plasmids; J. Yuan (Columbia University) for the plasmids SFB-ZRANB3, SFB-ZRANB3-Δ PIP and SFBZRANB3(Q519A); D. Ahel (Oxford University) for the plasmids YFP-ZRANB3 and Flag-ZRANB3; Z. Zhang (University of Delaware) for the plasmid pet15b-His5-PCNA; A. Grimson (Cornell University) for the shRNA plasmids for RNR-α, RNR-β and IRBIT. Research: Pershing Square Sohn Cancer Research Alliance grant (to Y.A.); Meyer Cancer Center grant (Weill Cornell Medicine) (to Y.A. and R.S.W.); and the Canadian Institutes of Health Research grant (MOP-82930) (to J.O.). Instrumentation and shared supplies: NIH DP2 New Innovator (1DP2GM114850); NSF CAREER (CHE-1351400); Office of Naval Research (ONR) Young Investigator (N00014-17-1-2529); Beckman Young Investigator; Sloan Fellowship (FG-2016-6379) (to Y.A.); Cornell NMR facility (NSF MRI: CHE-1531632; PI: Y.A.) and Cornell Imaging Center (NIH 1S10RR025502; PI: R.M. Williams).
Author information
Authors and Affiliations
Contributions
Y.F., M.J.C.L. and Y.A. designed the experiments. Y.F. and M.J.C.L. performed the experiments. S.W. synthesized ClF, CLA and FLU nucleotides. H.I. and J.O. performed electron microscopy analysis. I.M.E. assisted M.J.C.L. with targeted mutagenesis for binding-site analysis. M.J.C.L., T.M.P., J.C.B. and R.S.W. generated mouse embryonic fibroblast cultures. Y.F., M.J.C.L. and Y.A. analyzed and interpreted the data. Y.F., M.J.C.L. and Y.A. wrote the paper with proof-editing contributions from R.S.W. and J.O.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Supplementary Text and Figures
Supplementary Table 1–2, Supplementary Figures 1–42
Rights and permissions
About this article
Cite this article
Fu, Y., Long, M.J.C., Wisitpitthaya, S. et al. Nuclear RNR-α antagonizes cell proliferation by directly inhibiting ZRANB3. Nat Chem Biol 14, 943–954 (2018). https://doi.org/10.1038/s41589-018-0113-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-018-0113-5
This article is cited by
-
Wdr1 and cofilin are necessary mediators of immune-cell-specific apoptosis triggered by Tecfidera
Nature Communications (2021)