Abstract
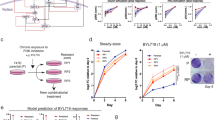
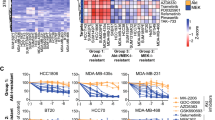
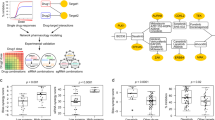
Dysregulation of the PI3K-AKT-mTOR signaling network is a prominent feature of breast cancers. However, clinical responses to drugs targeting this pathway have been modest, possibly because of dynamic changes in cellular signaling that drive resistance and limit drug efficacy. Using a quantitative chemoproteomics approach, we mapped kinome dynamics in response to inhibitors of this pathway and identified signaling changes that correlate with drug sensitivity. Maintenance of AURKA after drug treatment was associated with resistance in breast cancer models. Incomplete inhibition of AURKA was a common source of therapy failure, and combinations of PI3K, AKT or mTOR inhibitors with the AURKA inhibitor MLN8237 were highly synergistic and durably suppressed mTOR signaling, resulting in apoptosis and tumor regression in vivo. This signaling map identifies survival factors whose presence limits the efficacy of targeted therapies and reveals new drug combinations that may unlock the full potential of PI3K–AKT–mTOR pathway inhibitors in breast cancer.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70 (2012).
Janku, F. et al. Assessing PIK3CA and PTEN in early-phase trials with PI3K/AKT/mTOR inhibitors. Cell Rep 6, 377–387 (2014).
Dey, N., De, P. & Leyland-Jones, B. PI3K-AKT-mTOR inhibitors in breast cancers: from tumor cell signaling to clinical trials. Pharmacol. Ther. 175, 91–106 (2017).
O’Reilly, K. E. et al. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 66, 1500–1508 (2006).
Klempner, S. J., Myers, A. P. & Cantley, L. C. What a tangled web we weave: emerging resistance mechanisms to inhibition of the phosphoinositide 3-kinase pathway. Cancer Discov. 3, 1345–1354 (2013).
Shah, P.D. & Chandarlapaty, S. in PI3K-mTOR in Cancer and Cancer Therapy (eds. Dey, N., De, P. & Leyland-Jones, B.) 125–147 (Springer International Publishing, 2016).
Duncan, J. S. et al. Dynamic reprogramming of the kinome in response to targeted MEK inhibition in triple-negative breast cancer. Cell 149, 307–321 (2012).
Corcoran, R. B. et al. TORC1 suppression predicts responsiveness to RAF and MEK inhibition in BRAF-mutant melanoma. Sci. Transl. Med. 5, 196ra98 (2013).
Anderson, G. R. et al. PIK3CA mutations enable targeting of a breast tumor dependency through mTOR-mediated MCL-1 translation. Sci. Transl. Med. 8, 369ra175 (2016).
Boussemart, L. et al. eIF4F is a nexus of resistance to anti-BRAF and anti-MEK cancer therapies. Nature 513, 105–109 (2014).
Elkabets, M. et al. mTORC1 inhibition is required for sensitivity to PI3K p110α inhibitors in PIK3CA-mutant breast cancer. Sci. Transl. Med. 5, 196ra99 (2013).
Vora, S. R. et al. CDK 4/6 inhibitors sensitize PIK3CA mutant breast cancer to PI3K inhibitors. Cancer Cell 26, 136–149 (2014).
Bantscheff, M. et al. Quantitative chemical proteomics reveals mechanisms of action of clinical ABL kinase inhibitors. Nat. Biotechnol. 25, 1035–1044 (2007).
Sos, M. L. et al. Oncogene mimicry as a mechanism of primary resistance to BRAF inhibitors. Cell Rep. 8, 1037–1048 (2014).
Stuhlmiller, T. J. et al. Inhibition of lapatinib-induced kinome reprogramming in ERBB2-positive breast cancer by targeting BET family bromodomains. Cell Rep 11, 390–404 (2015).
Lens, S. M. A., Voest, E. E. & Medema, R. H. Shared and separate functions of polo-like kinases and aurora kinases in cancer. Nat. Rev. Cancer 10, 825–841 (2010).
Bosch, A. et al. PI3K inhibition results in enhanced estrogen receptor function and dependence in hormone receptor–positive breast cancer. Sci. Transl. Med. 7, 283ra51 (2015).
Leroy, C. et al. Activation of IGF1R/p110β/AKT/mTOR confers resistance to α-specific PI3K inhibition. Breast Cancer Res. 18, 41 (2016).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550 (2005).
Foucquier, J. & Guedj, M. Analysis of drug combinations: current methodological landscape. Pharmacol. Res. Perspect. 3, e00149 (2015).
Fruman, D. A. & Rommel, C. PI3K and cancer: lessons, challenges and opportunities. Nat. Rev. Drug Discov. 13, 140–156 (2014).
Nikonova, A. S., Astsaturov, I., Serebriiskii, I. G., Dunbrack, R. L. Jr. & Golemis, E. A. Aurora A kinase (AURKA) in normal and pathological cell division. Cell. Mol. Life Sci. 70, 661–687 (2013).
Piccart, M. et al. Everolimus plus exemestane for hormone-receptor-positive, human epidermal growth factor receptor-2-negative advanced breast cancer: overall survival results from BOLERO-2†. Ann. Oncol 25, 2357–2362 (2014).
Choo, A. Y., Yoon, S.-O., Kim, S. G., Roux, P. P. & Blenis, J. Rapamycin differentially inhibits S6Ks and 4E-BP1 to mediate cell-type-specific repression of mRNA translation. Proc. Natl. Acad. Sci. USA 105, 17414–17419 (2008).
Vadlakonda, L., Dash, A., Pasupuleti, M., Anil Kumar, K. & Reddanna, P. The paradox of Akt-mTOR interactions. Front. Oncol. 3, 165 (2013).
Laplante, M. & Sabatini, D. M. mTOR signaling in growth control and disease. Cell 149, 274–293 (2012).
Volkmann, N., Marassi, F. M., Newmeyer, D. D. & Hanein, D. The rheostat in the membrane: BCL-2 family proteins and apoptosis. Cell Death Differ. 21, 206–215 (2014).
West, M. J., Stoneley, M. & Willis, A. E. Translational induction of the c-myc oncogene via activation of the FRAP/TOR signalling pathway. Oncogene 17, 769–780 (1998).
Sears, R. et al. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev 14, 2501–2514 (2000).
Diehl, J. A., Cheng, M., Roussel, M. F. & Sherr, C. J. Glycogen synthase kinase-3β regulates cyclin D1 proteolysis and subcellular localization. Genes Dev. 12, 3499–3511 (1998).
Yang, H., He, L., Kruk, P., Nicosia, S. V. & Cheng, J. Q. Aurora-A induces cell survival and chemoresistance by activation of Akt through a p53-dependent manner in ovarian cancer cells. Int. J. Cancer 119, 2304–2312 (2006).
Dar, A. A., Belkhiri, A. & El-Rifai, W. The aurora kinase A regulates GSK-3β in gastric cancer cells. Oncogene 28, 866–875 (2009).
Martins, M. M. et al. Linking tumor mutations to drug responses via a quantitative chemical-genetic interaction map. Cancer Discov. 5, 154–167 (2015).
Muellner, M. K. et al. A chemical-genetic screen reveals a mechanism of resistance to PI3K inhibitors in cancer. Nat. Chem. Biol. 7, 787–793 (2011).
Ilic, N., Utermark, T., Widlund, H. R. & Roberts, T. M. PI3K-targeted therapy can be evaded by gene amplification along the MYC-eukaryotic translation initiation factor 4E (eIF4E) axis. Proc. Natl. Acad. Sci. USA 108, E699–E708 (2011).
Liu, P. et al. Oncogenic PIK3CA-driven mammary tumors frequently recur via PI3K pathway-dependent and PI3K pathway-independent mechanisms. Nat. Med. 17, 1116–1120 (2011).
Nomanbhoy, T. K. et al. Chemoproteomic evaluation of target engagement by the cyclin-dependent kinase 4 and 6 inhibitor palbociclib correlates with cancer cell response. Biochemistry 55, 5434–5441 (2016).
Roux, P. P. et al. RAS/ERK signaling promotes site-specific ribosomal protein S6 phosphorylation via RSK and stimulates cap-dependent translation. J. Biol. Chem. 282, 14056–14064 (2007).
She, Q.-B. et al. 4E-BP1 is a key effector of the oncogenic activation of the AKT and ERK signaling pathways that integrates their function in tumors. Cancer Cell 18, 39–51 (2010).
den Hollander, J. et al. Aurora kinases A and B are up-regulated by Myc and are essential for maintenance of the malignant state. Blood 116, 1498–1505 (2010).
Lu, L. et al. Aurora kinase A mediates c-Myc’s oncogenic effects in hepatocellular carcinoma. Mol. Carcinog. 54, 1467–1479 (2015).
Zheng, F. et al. Nuclear AURKA acquires kinase-independent transactivating function to enhance breast cancer stem cell phenotype. Nat. Commun. 7, 10180 (2016).
Ferrell, J. E. Jr. Self-perpetuating states in signal transduction: positive feedback, double-negative feedback and bistability. Curr. Opin. Cell Biol. 14, 140–148 (2002).
Katsha, A. et al. Activation of EIF4E by aurora kinase A depicts a novel druggable axis in everolimus-resistant cancer cells. Clin. Cancer Res. 23, 3756–3768 (2017).
Brewer Savannah, K. J. et al. Dual targeting of mTOR and aurora-A kinase for the treatment of uterine Leiomyosarcoma. Clin. Cancer Res. 18, 4633–4645 (2012).
Liu, L.-L. et al. Inhibition of mTOR pathway sensitizes acute myeloid leukemia cells to aurora inhibitors by suppression of glycolytic metabolism. Mol. Cancer Res. 11, 1326–1336 (2013).
Baselga, J. et al. Buparlisib plus fulvestrant versus placebo plus fulvestrant in postmenopausal, hormone receptor-positive, HER2-negative, advanced breast cancer (BELLE-2): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 18, 904–916 (2017).
Zwang, Y. et al. Synergistic interactions with PI3K inhibition that induce apoptosis. eLife 6, e24523 (2017).
Barr, P. M. et al. Phase II intergroup trial of alisertib in relapsed and refractory peripheral T-cell lymphoma and transformed mycosis fungoides: SWOG 1108. J. Clin. Oncol. Clin. Oncol. 33, 2399–2404 (2015).
Neve, R. M. et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 10, 515–527 (2006).
Barvian, M. et al. Pyrido[2,3-d]pyrimidin-7-one inhibitors of cyclin-dependent kinases. J. Med. Chem. 43, 4606–4616 (2000).
Daub, H. et al. Kinase-selective enrichment enables quantitative phosphoproteomics of the kinome across the cell cycle. Mol. Cell 31, 438–448 (2008).
Blake, J. F. et al. Discovery of pyrrolopyrimidine inhibitors of Akt. Bioorg. Med. Chem. Lett. 20, 5607–5612 (2010).
Tanaka, M. et al. An unbiased cell morphology-based screen for new, biologically active small molecules. PLoS Biol. 3, e128 (2005).
Bankston, D. et al. A scaleable synthesis of BAY 43–9006: a potent Raf kinase inhibitor for the treatment of cancer. Org. Process Res. Dev. 6, 777–781 (2002).
Statsuk, A. V. et al. Tuning a three-component reaction for trapping kinase substrate complexes. J. Am. Chem. Soc. 130, 17568–17574 (2008).
MacLean, B. et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968 (2010).
Choi, M. et al. MSstats: an R package for statistical analysis of quantitative mass spectrometry-based proteomic experiments. Bioinformatics 30, 2524–2526 (2014).
Lehár, J. et al. Synergistic drug combinations tend to improve therapeutically relevant selectivity. Nat. Biotechnol. 27, 659–666 (2009).
Acknowledgements
The authors would like to thank members of the Bandyopadhyay laboratory for helpful discussions and technical assistance. We also thank A. Beardsley, E. Markegard, D. Ruggero and W. Weiss for helpful discussions and reagents. This work was supported in part by NCI U01CA168370 (S.B.), NIGMS R01GM107671 (S.B.), NCI R01CA170447 (A.G.), Prospect Creek Foundation (S.B., A.G.), OHSU Pilot Project Funding (S.B., J.K.), American Cancer Society Postdoctoral Fellowship (J.D.G.), the Gazarian Foundation (A.G.), DOD W81XWH-12-1-0272 and DOD W81XWH-16-1-0603 (A.G.).
Author information
Authors and Affiliations
Contributions
H.J.D., J.T.W., K.M.S, J.K., J.D.G., and S.B. contributed toward study conceptualization. H.J.D., J.T.W., and J.D.G. performed data analyses supporting the study. H.J.D. designed and performed the majority of experiments. N.B. assisted with samples for initial MIBs/MS profiling. R.S.L. J.T.W., and J.D.G. provided technical advice and guided the interpretation of mass spectrometry data from MIB/MS profiling. R.C. and O.M. assisted with animal studies, and K.N.S. helped with additional experiments. H.J.D. and S.B. composed the original draft, and all authors contributed to manuscript finalization. S.B. and A.G. supervised the study.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–19
Supplementary Dataset 1
Kinase activity scores and P values from MIBs/MS profiling.
Supplementary Dataset 2
Gene set enrichment analysis (GSEA) of BYL719-treated cells.
Supplementary Dataset 3
Receptor status, synergy scores (Loewe and Bliss) and assessment of increase in apoptosis over single-agent or combinations tested in this study.
Supplementary Dataset 4
MYC signature derived from isogenic MCF10A cell lines.
Rights and permissions
About this article
Cite this article
Donnella, H.J., Webber, J.T., Levin, R.S. et al. Kinome rewiring reveals AURKA limits PI3K-pathway inhibitor efficacy in breast cancer. Nat Chem Biol 14, 768–777 (2018). https://doi.org/10.1038/s41589-018-0081-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-018-0081-9
This article is cited by
-
Predictive, preventive, and personalized medicine in breast cancer: targeting the PI3K pathway
Journal of Translational Medicine (2024)
-
Deletion of Aurora kinase A prevents the development of polycystic kidney disease in mice
Nature Communications (2024)
-
NCAPH serves as a prognostic factor and promotes the tumor progression in glioma through PI3K/AKT signaling pathway
Molecular and Cellular Biochemistry (2024)
-
HMMR promotes prostate cancer proliferation and metastasis via AURKA/mTORC2/E2F1 positive feedback loop
Cell Death Discovery (2023)
-
Identification, characterization, and prognosis investigation of pivotal genes shared in different stages of breast cancer
Scientific Reports (2023)