Abstract
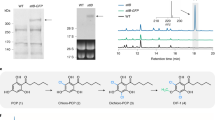
The ultimate step in the formation of thebaine, a pentacyclic opiate alkaloid readily converted to the narcotic analgesics codeine and morphine in the opium poppy, has long been presumed to be a spontaneous reaction. We have detected and purified a novel enzyme from opium poppy latex that is capable of the efficient formation of thebaine from (7S)-salutaridinol 7-O-acetate at the expense of labile hydroxylated byproducts, which are preferentially produced by spontaneous allylic elimination. Remarkably, thebaine synthase (THS), a member of the pathogenesis-related 10 protein (PR10) superfamily, is encoded within a novel gene cluster in the opium poppy genome that also includes genes encoding the four biosynthetic enzymes immediately upstream. THS is a missing component that is crucial to the development of fermentation-based opiate production and dramatically improves thebaine yield in engineered yeast.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Ehrenworth, A. M. & Peralta-Yahya, P. Accelerating the semisynthesis of alkaloid-based drugs through metabolic engineering. Nat. Chem. Biol. 13, 249–258 (2017).
Tatsis, E. C. & O’Connor, S. E. New developments in engineering plant metabolic pathways. Curr. Opin. Biotechnol. 42, 126–132 (2016).
Galanie, S., Thodey, K., Trenchard, I. J., Filsinger Interrante, M. & Smolke, C. D. Complete biosynthesis of opioids in yeast. Science 349, 1095–1100 (2015).
Winzer, T. et al. Plant science. Morphinan biosynthesis in opium poppy requires a P450-oxidoreductase fusion protein. Science 349, 309–312 (2015).
Farrow, S. C., Hagel, J. M., Beaudoin, G. A. W., Burns, D. C. & Facchini, P. J. Stereochemical inversion of (S)-reticuline by a cytochrome P450 fusion in opium poppy. Nat. Chem. Biol. 11, 728–732 (2015).
Service, R.F. Final step in sugar-to-morphine conversion deciphered. Science https://doi.org/10.1126/science.aac6897/ (2015).
Keener, A.B. Yeast-based opioid production completed. The Scientist https://www.the-scientist.com/?articles.view/articleNo/43739/title/Yeast-Based-Opioid-Production-Completed/ (2015).
Battersby, A. R. Tilden Lecture: The biosynthesis of alkaloids. Proc. Chem. Soc. 0, 189–228 (1963).
Lenz, R. & Zenk, M. H. Closure of the oxide bridge in morphine biosynthesis. Tetrahedr. Lett. 35, 3897–3900 (1994).
Lenz, R. & Zenk, M. H. Acetyl coenzyme A:salutaridinol-7-O-acetyltransferase from Papaver somniferum plant cell cultures. The enzyme catalyzing the formation of thebaine in morphine biosynthesis. J. Biol. Chem. 270, 31091–31096 (1995).
Barton, D. H. R., Bhakuni, D. S., James, R. & Kirby, G. W. Phenol oxidation and biosynthesis. Part XII. Stereochemical studies related to the biosynthesis of the morphine alkaloids. J. Chem. Soc. 0, 128–132 (1967).
Lotter, H., Gollwitzer, J. & Zenk, M. H. Revision of the configuration at C-7 of salutaridinol-I, the natural intermediate in morphine biosynthesis. Tetrahedr. Lett. 33, 2443–2446 (1992).
Hagel, J. M. & Facchini, P. J. Benzylisoquinoline alkaloid metabolism: a century of discovery and a brave new world. Plant Cell Physiol. 54, 647–672 (2013).
Ziegler, J. & Facchini, P. J. Alkaloid biosynthesis: metabolism and trafficking. Annu. Rev. Plant Biol. 59, 735–769 (2008).
Fisinger, U., Grobe, N. & Zenk, M. H. Thebaine synthase: a new enzyme in the morphine pathway in Papaver somniferum. Nat. Prod. Commun. 2, 249–253 (2007).
Barton, D. H. R. et al. Investigations on the biosynthesis of morphine alkaloids. J. Chem. Soc. 65, 2423–2438 (1965).
Raith, K. et al. Electrospray tandem mass spectrometric investigations of morphinans. J. Am. Soc. Mass Spectrom. 14, 1262–1269 (2003).
Pennanen, K., Kotiaho, T., Huikko, K. & Kostiainen, R. Identification of ozone-oxidation products of oxycodone by electrospray ion trap mass spectrometry. J. Mass Spectrom. 36, 791–797 (2001).
Sabarna, K. Approaches to isolating a cDNA encoding thebaine synthase of morphine biosynthesis from opium poppy Papaver somniferum. PhD thesis, Martin Luther University Halle-Wittenberg (2006).
Fernandes, H., Michalska, K., Sikorski, M. & Jaskolski, M. Structural and functional aspects of PR-10 proteins. FEBS J. 280, 1169–1199 (2013).
Lee, E.-J. & Facchini, P. Norcoclaurine synthase is a member of the pathogenesis-related 10/Bet v1 protein family. Plant Cell 22, 3489–3503 (2010).
Mikkelsen, M. D. et al. Microbial production of indolylglucosinolate through engineering of a multi-gene pathway in a versatile yeast expression platform. Metab. Eng. 14, 104–111 (2012).
Desgagné-Penix, I., Farrow, S. C., Cram, D., Nowak, J. & Facchini, P. J. Integration of deep transcript and targeted metabolite profiles for eight cultivars of opium poppy. Plant Mol. Biol. 79, 295–313 (2012).
Dang, T. T., Onoyovwi, A., Farrow, S. C. & Facchini, P. J. Biochemical genomics for gene discovery in benzylisoquinoline alkaloid biosynthesis in opium poppy and related species. Methods Enzymol. 515, 231–266 (2012).
Boycheva, S., Daviet, L., Wolfender, J.-L. & Fitzpatrick, T. B. The rise of operon-like gene clusters in plants. Trends Plant Sci. 19, 447–459 (2014).
Nützmann, H.-W. & Osbourn, A. Gene clustering in plant specialized metabolism. Curr. Opin. Biotechnol. 26, 91–99 (2014).
Winzer, T. et al. A Papaver somniferum 10-gene cluster for synthesis of the anticancer alkaloid noscapine. Science 336, 1704–1708 (2012).
Ilari, A. et al. Structural basis of enzymatic (S)-norcoclaurine biosynthesis. J. Biol. Chem. 284, 897–904 (2009).
Robin, A. Y., Giustini, C., Graindorge, M., Matringe, M. & Dumas, R. Crystal structure of norcoclaurine-6-O-methyltransferase, a key rate-limiting step in the synthesis of benzylisoquinoline alkaloids. Plant J. 87, 641–653 (2016).
Löbermann, F., Mayer, P. & Trauner, D. Biomimetic synthesis of (-)-pycnanthuquinone C through the Diels-Alder reaction of a vinyl quinone. Angew. Chem. Int. Ed. Engl. 49, 6199–6202 (2010).
Fossati, E., Narcross, L., Ekins, A., Falgueyret, J. P. & Martin, V. J. Synthesis of morphinan alkaloids in Saccharomyces cerevisiae. PLoS One 10, e0124459 (2015).
Nessler, C. L., Allen, R. D. & Galewsky, S. Identification and characterization of latex-specific proteins in opium poppy. Plant Physiol. 79, 499–504 (1985).
Bieniossek, C. et al. Automated unrestricted multigene recombineering for multiprotein complex production. Nat. Methods 6, 447–450 (2009).
Farrow, S. C., Hagel, J. M. & Facchini, P. J. Transcript and metabolite profiling in cell cultures of 18 plant species that produce benzylisoquinoline alkaloids. Phytochemistry 77, 79–88 (2012).
Chang, L., Hagel, J. M. & Facchini, P. J. Isolation and characterization of O-methyltransferases involved in the biosynthesis of glaucine in Glaucium flavum. Plant Physiol. 169, 1127–1140 (2015).
Morris, J. S. et al. Plug-and-play benzylisoquinoline alkaloid biosynthetic gene discovery in engineered yeast. Methods Enzymol. 575, 143–178 (2016).
Schmidt, J., Boettcher, C., Kuhnt, C., Kutchan, T. M. & Zenk, M. H. Poppy alkaloid profiling by electrospray tandem mass spectrometry and electrospray FT-ICR mass spectrometry after [ring-13C6]-tyramine feeding. Phytochemistry 68, 189–202 (2007).
Gambino, G., Perrone, I. & Gribaudo, I. A rapid and effective method for RNA extraction from different tissues of grapevine and other woody plants. Phytochem. Anal. 19, 520–525 (2008).
Acknowledgements
We are grateful to L. Brechenmacher and the Southern Alberta Mass Spectrometry Centre for assistance with the proteomics analysis, S. Yeaman for guidance with the genome assembly, and T. Back for advice on chemical reaction mechanisms. We acknowledge the expert services provided by the McGill University-Genome Québec Innovation Centre with respect to genome sequencing and preliminary assembly. We also thank A. Pigula, S. Muley, E. Eberhard, A. Kumar, K. Hetenyi, I. Esaid, H. Tang, H. Roth, M. Schmalisch, L. Hom, C. Savile, P. Seufer-Wasserthal, T. Noh, B. Walsh, and R. J. Kirk for technical assistance and project guidance. This work was supported by Genopaver, LLC., Epimeron Inc. and funds awarded through the Industrial Research Assistance Program (IRAP; Project 86155) operated by the National Research Council of Canada to Epimeron, Inc.
Author information
Authors and Affiliations
Contributions
X.C., J.M.H., J.E.T., G.C., G.M.V., and P.J.F. contributed to the project design; X.C. performed most of the enzymology, protein purification, molecular biology, gene silencing, and transcript profiling work; J.M.H. conducted the high-resolution LC-MS analysis and proposed the reaction mechanism; L.C., S.A.S., and M.E.-N. developed testing conditions and characterized gene candidates in yeast; R.E. and J.C. designed and built plasmids and yeast strains; Y.Y., H.-Y.C., and A.B.I. developed and performed LC-MS analysis for yeast fermentation experiments; X.C., J.M.H., and P.J.F. wrote the manuscript. P.J.F. supervised the project. All authors have read and approved the content of the manuscript.
Corresponding author
Ethics declarations
Competing interests
Patent applications related to this work have been filed (PCT/CA2017/050779 and PCT/US2017/039589).
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–22, Supplementary Tables 1–8, Supplementary Note 1–3
Rights and permissions
About this article
Cite this article
Chen, X., Hagel, J.M., Chang, L. et al. A pathogenesis-related 10 protein catalyzes the final step in thebaine biosynthesis. Nat Chem Biol 14, 738–743 (2018). https://doi.org/10.1038/s41589-018-0059-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-018-0059-7
This article is cited by
-
Elucidation of the (R)-enantiospecific benzylisoquinoline alkaloid biosynthetic pathways in sacred lotus (Nelumbo nucifera)
Scientific Reports (2023)
-
Emerging functions within the enzyme families of plant alkaloid biosynthesis
Phytochemistry Reviews (2023)
-
Alkaloid binding to opium poppy major latex proteins triggers structural modification and functional aggregation
Nature Communications (2022)
-
Insights into opium poppy (Papaver spp.) genetic diversity from genotyping-by-sequencing analysis
Scientific Reports (2022)
-
A functionally conserved STORR gene fusion in Papaver species that diverged 16.8 million years ago
Nature Communications (2022)