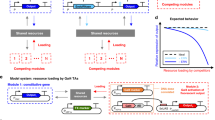
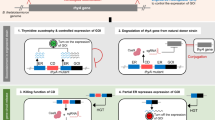
Abstract
The increasing use of engineered organisms for industrial, clinical, and environmental applications poses a growing risk of spreading hazardous biological entities into the environment. To address this biosafety issue, significant effort has been invested in creating ways to confine these organisms and transgenic materials. Emerging technologies in synthetic biology involving genetic circuit engineering, genome editing, and gene expression regulation have led to the development of novel biocontainment systems. In this perspective, we highlight recent advances in biocontainment and suggest a number of approaches for future development, which may be applied to overcome remaining challenges in safeguard implementation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Khalil, A. S. & Collins, J. J. Synthetic biology: applications come of age. Nat. Rev. Genet. 11, 367–379 (2010).
Lee, J. W. et al. Systems metabolic engineering of microorganisms for natural and non-natural chemicals. Nat. Chem. Biol. 8, 536–546 (2012).
Cameron, D. E., Bashor, C. J. & Collins, J. J. A brief history of synthetic biology. Nat. Rev. Microbiol. 12, 381–390 (2014).
Berg, P., Baltimore, D., Brenner, S., Roblin, R. O. & Singer, M. F. Summary statement of the Asilomar conference on recombinant DNA molecules. Proc. Natl Acad. Sci. USA 72, 1981–1984 (1975).
Berg, P., Baltimore, D., Brenner, S., Roblin, R. O. III & Singer, M. F. Asilomar conference on recombinant DNA molecules. Science 188, 991–994 (1975).
Wilson, D. J. NIH guidelines for research involving recombinant DNA molecules. Account. Res. 3, 177–185 (1993).
Sears, M. K. et al. Impact of Bt corn pollen on monarch butterfly populations: a risk assessment. Proc. Natl Acad. Sci. USA 98, 11937–11942 (2001).
Stanley-Horn, D. E. et al. Assessing the impact of Cry1Ab-expressing corn pollen on monarch butterfly larvae in field studies. Proc. Natl Acad. Sci. USA 98, 11931–11936 (2001).
Snow, A. A. Transgenic crops why gene flow matters. Nat. Biotechnol. 20, 542 (2002).
Giovannetti, M. The ecological risks of transgenic plants. Riv. Biol. 96, 207–223 (2003).
Hills, M. J., Hall, L., Arnison, P. G. & Good, A. G. Genetic use restriction technologies (GURTs): strategies to impede transgene movement. Trends Plant Sci. 12, 177–183 (2007).
Quist, D. & Chapela, I. H. Transgenic DNA introgressed into traditional maize landraces in Oaxaca, Mexico. Nature 414, 541–543 (2001).
Colomer-Lluch, M., Imamovic, L., Jofre, J. & Muniesa, M. Bacteriophages carrying antibiotic resistance genes in fecal waste from cattle, pigs, and poultry. Antimicrob. Agents Chemother. 55, 4908–4911 (2011).
Moe-Behrens, G. H., Davis, R. & Haynes, K. A. Preparing synthetic biology for the world. Front. Microbiol. 4, 5 (2013). This review provides a detailed summary of the development of pioneering biocontainment systems.
Dana, G. V., Kuiken, T., Rejeski, D. & Snow, A. A. Synthetic biology: four steps to avoid a synthetic-biology disaster. Nature 483, 29 (2012).
Wright, O., Stan, G. B. & Ellis, T. Building-in biosafety for synthetic biology. Microbiology 159, 1221–1235 (2013).
Schmidt, M. & de Lorenzo, V. Synthetic bugs on the loose: containment options for deeply engineered (micro)organisms. Curr. Opin. Biotechnol. 38, 90–96 (2016).
Schmidt, M. & Pei, L. in Hydrocarbon and Lipid Microbiology Protocols: Synthetic and Systems Biology - Tools (eds. McGenity, T.J., Timmis, K.N. & Nogales, B.) 185–199 (Springer Berlin Heidelberg, Berlin, Heidelberg, 2016).
Oliver, M. J., Quisenberry, J. E., Trolinder, N. L. G. & Keim, D. H. Control of plant gene expression. US Patent 5,723,765 (1998).
Naldini, L. et al. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272, 263–267 (1996).
Dull, T. et al. A third-generation lentivirus vector with a conditional packaging system. J. Virol. 72, 8463–8471 (1998).
Burns, J. C., Friedmann, T., Driever, W., Burrascano, M. & Yee, J. K. Vesicular stomatitis virus G glycoprotein pseudotyped retroviral vectors: concentration to very high titer and efficient gene transfer into mammalian and nonmammalian cells. Proc. Natl Acad. Sci. USA 90, 8033–8037 (1993).
Steidler, L. et al. Biological containment of genetically modified Lactococcus lactis for intestinal delivery of human interleukin 10. Nat. Biotechnol. 21, 785–789 (2003). This article describes one of the earliest clinical studies involving a probiotic strain equipped with a biocontainment system.
Bahey-El-Din, M., Casey, P. G., Griffin, B. T. & Gahan, C. G. Efficacy of a Lactococcus lactis ΔpyrG vaccine delivery platform expressing chromosomally integrated hly from Listeria monocytogenes. Bioeng. Bugs 1, 66–74 (2010).
Ronchel, M. C. & Ramos, J. L. Dual system to reinforce biological containment of recombinant bacteria designed for rhizoremediation. Appl. Environ. Microbiol. 67, 2649–2656 (2001).
Lajoie, M. J. et al. Genomically recoded organisms expand biological functions. Science 342, 357–360 (2013).
Rovner, A. J. et al. Recoded organisms engineered to depend on synthetic amino acids. Nature 518, 89–93 (2015). This paper (along with the one in ref. 28) presents the development of synthetic auxotroph-based biocontainment systems requiring an unnatural amino acid for the synthesis of essential proteins, in two parallel studies.
Mandell, D. J. et al. Biocontainment of genetically modified organisms by synthetic protein design. Nature 518, 55–60 (2015).
Callura, J. M., Dwyer, D. J., Isaacs, F. J., Cantor, C. R. & Collins, J. J. Tracking, tuning, and terminating microbial physiology using synthetic riboregulators. Proc. Natl Acad Sci. USA 107, 15898–15903 (2010).
Gallagher, R. R., Patel, J. R., Interiano, A. L., Rovner, A. J. & Isaacs, F. J. Multilayered genetic safeguards limit growth of microorganisms to defined environments. Nucleic Acids Res. 43, 1945–1954 (2015).
Cai, Y. et al. Intrinsic biocontainment: multiplex genome safeguards combine transcriptional and recombinational control of essential yeast genes. Proc. Natl Acad. Sci. USA 112, 1803–1808 (2015).
Agmon, N. et al. Low escape-rate genome safeguards with minimal molecular perturbation of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA 114, E1470–E1479 (2017).
Huang, S. et al. Coupling spatial segregation with synthetic circuits to control bacterial survival. Mol. Syst. Biol. 12, 859 (2016).
Chan, C. T., Lee, J. W., Cameron, D. E., Bashor, C. J. & Collins, J. J. ‘Deadman’ and ‘Passcode’ microbial kill switches for bacterial containment. Nat. Chem. Biol. 12, 82–86 (2016). This article describes one of the first safeguard systems designed to provide programmable conditions for biocontainment.
Molina, L., Ramos, C., Ronchel, M. C., Molin, S. & Ramos, J. L. Construction of an efficient biologically contained pseudomonas putida strain and its survival in outdoor assays. Appl. Environ. Microbiol. 64, 2072–2078 (1998).
Contreras, A., Molin, S. & Ramos, J. L. Conditional-suicide containment system for bacteria which mineralize aromatics. Appl. Environ. Microbiol. 57, 1504–1508 (1991).
Weaver, K. E. The par toxin-antitoxin system from Enterococcus faecalis plasmid pAD1 and its chromosomal homologs. RNA Biol. 9, 1498–1503 (2012).
Durand, S., Jahn, N., Condon, C. & Brantl, S. Type I toxin-antitoxin systems in Bacillus subtilis. RNA Biol. 9, 1491–1497 (2012).
Diago-Navarro, E. et al. parD toxin-antitoxin system of plasmid R1–basic contributions, biotechnological applications and relationships with closely-related toxin-antitoxin systems. FEBS J. 277, 3097–3117 (2010).
Yagura, M., Nishio, S. Y., Kurozumi, H., Wang, C. F. & Itoh, T. Anatomy of the replication origin of plasmid ColE2-P9. J. Bacteriol. 188, 999–1010 (2006).
del Solar, G., Giraldo, R., Ruiz-Echevarría, M. J., Espinosa, M. & Díaz-Orejas, R. Replication and control of circular bacterial plasmids. Microbiol. Mol. Biol. Rev. 62, 434–464 (1998).
Caliando, B. J. & Voigt, C. A. Targeted DNA degradation using a CRISPR device stably carried in the host genome. Nat. Commun. 6, 6989 (2015).
Chavez, A. et al. Precise Cas9 targeting enables genomic mutation prevention. Proc. Natl Acad. Sci. USA https://doi.org/10.1073/pnas.1718148115 (2018).
Callaway, E. ‘Minimal’ cell raises stakes in race to harness synthetic life. Nature 531, 557–558 (2016).
Johns, N. I., Blazejewski, T., Gomes, A. L. & Wang, H. H. Principles for designing synthetic microbial communities. Curr. Opin. Microbiol. 31, 146–153 (2016).
Mee, M. T., Collins, J. J., Church, G. M. & Wang, H. H. Syntrophic exchange in synthetic microbial communities. Proc. Natl Acad. Sci. USA 111, E2149–E2156 (2014).
Wang, K., Neumann, H., Peak-Chew, S. Y. & Chin, J. W. Evolved orthogonal ribosomes enhance the efficiency of synthetic genetic code expansion. Nat. Biotechnol. 25, 770–777 (2007).
Neumann, H., Wang, K., Davis, L., Garcia-Alai, M. & Chin, J. W. Encoding multiple unnatural amino acids via evolution of a quadruplet-decoding ribosome. Nature 464, 441–444 (2010).
Terasaka, N., Hayashi, G., Katoh, T. & Suga, H. An orthogonal ribosome-tRNA pair via engineering of the peptidyl transferase center. Nat. Chem. Biol. 10, 555–557 (2014).
Soye, B. J. D., Patel, J. R., Isaacs, F. J. & Jewett, M. C. Repurposing the translation apparatus for synthetic biology. Curr. Opin. Chem. Biol. 28, 83–90 (2015).
Marlière, P. et al. Chemical evolution of a bacterium’s genome. Angew. Chem. Int. Edn. Engl. 50, 7109–7114 (2011). This article involved the engineering of a bacterial strain that is capable of using 5-chlorouracil to replace thymine in DNA, which is one of the first examples of a xenobiological system.
Pinheiro, V. B. et al. Synthetic genetic polymers capable of heredity and evolution. Science 336, 341–344 (2012). This paper presents the development of xenobiological systems that replace canonical nucleosides with different types of synthetic analogs.
Malyshev, D. A. et al. A semi-synthetic organism with an expanded genetic alphabet. Nature 509, 385–388 (2014).
Adamala, K. P., Martin-Alarcon, D. A., Guthrie-Honea, K. R. & Boyden, E. S. Engineering genetic circuit interactions within and between synthetic minimal cells. Nat. Chem. 9, 431–439 (2017). This article describes the capability of a cell-free system to replace living entities in a range of applications in biomedicine and biotechnology.
Pardee, K. et al. Paper-based synthetic gene networks. Cell 159, 940–954 (2014).
Pardee, K. et al. Rapid, low-cost detection of Zika virus using programmable biomolecular components. Cell 165, 1255–1266 (2016).
Pardee, K. et al. Portable, on-demand biomolecular manufacturing. Cell 167, 248–259.e12 (2016).
Dudley, Q. M., Karim, A. S. & Jewett, M. C. Cell-free metabolic engineering: biomanufacturing beyond the cell. Biotechnol. J. 10, 69–82 (2015).
Garamella, J., Marshall, R., Rustad, M. & Noireaux, V. The All E. coli TX-TL Toolbox 2.0: A platform for cell-free synthetic biology. ACS Synth. Biol. 5, 344–355 (2016).
Hong, S. H. et al. Cell-free protein synthesis from a release factor 1 deficient Escherichia coli activates efficient and multiple site-specific nonstandard amino acid incorporation. ACS Synth. Biol. 3, 398–409 (2014).
Lu, T. K. & Koeris, M. S. The next generation of bacteriophage therapy. Curr. Opin. Microbiol. 14, 524–531 (2011).
Krom, R. J., Bhargava, P., Lobritz, M. A. & Collins, J. J. Engineered phagemids for nonlytic, targeted antibacterial therapies. Nano. Lett. 15, 4808–4813 (2015).
Bikard, D. et al. Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nat. Biotechnol. 32, 1146–1150 (2014).
Citorik, R. J., Mimee, M. & Lu, T. K. Sequence-specific antimicrobials using efficiently delivered RNA-guided nucleases. Nat. Biotechnol. 32, 1141–1145 (2014).
Ando, H., Lemire, S., Pires, D. P. & Lu, T. K. Engineering modular viral scaffolds for targeted bacterial population editing. Cell Syst. 1, 187–196 (2015).
Yosef, I., Manor, M., Kiro, R. & Qimron, U. Temperate and lytic bacteriophages programmed to sensitize and kill antibiotic-resistant bacteria. Proc. Natl Acad. Sci. USA 112, 7267–7272 (2015).
Champer, J., Buchman, A. & Akbari, O. S. Cheating evolution: engineering gene drives to manipulate the fate of wild populations. Nat. Rev. Genet. 17, 146–159 (2016).
DiCarlo, J. E., Chavez, A., Dietz, S. L., Esvelt, K. M. & Church, G. M. Safeguarding CRISPR-Cas9 gene drives in yeast. Nat. Biotechnol. 33, 1250–1255 (2015).
Gantz, V. M. & Bier, E. Genome editing. The mutagenic chain reaction: a method for converting heterozygous to homozygous mutations. Science 348, 442–444 (2015).
Gantz, V. M. et al. Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proc. Natl Acad. Sci. USA 112, E6736–E6743 (2015).
Hammond, A. et al. A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae. Nat. Biotechnol. 34, 78–83 (2016).
Wright, O., Delmans, M., Stan, G. B. & Ellis, T. GeneGuard: a modular plasmid system designed for biosafety. ACS Synth. Biol. 4, 307–316 (2015).
Molin, S. et al. Conditional suicide system for containment of bacteria and plasmids. Nat. Biotechnol. 5, 1315–1318 (1987).
Bej, A. K., Perlin, M. H. & Atlas, R. M. Model suicide vector for containment of genetically engineered microorganisms. Appl. Environ. Microbiol. 54, 2472–2477 (1988).
Knudsen, S. M. & Karlström, O. H. Development of efficient suicide mechanisms for biological containment of bacteria. Appl. Environ. Microbiol. 57, 85–92 (1991).
Poulsen, L. K., Larsen, N. W., Molin, S. & Andersson, P. A family of genes encoding a cell-killing function may be conserved in all gram-negative bacteria. Mol. Microbiol. 3, 1463–1472 (1989).
Braat, H. et al. A phase I trial with transgenic bacteria expressing interleukin-10 in Crohn’s disease. Clin. Gastroenterol. Hepatol. 4, 754–759 (2006).
Meinhardt, S. et al. Novel insights from hybrid LacI/GalR proteins: family-wide functional attributes and biologically significant variation in transcription repression. Nucleic Acids Res. 40, 11139–11154 (2012).
Meinhardt, S. & Swint-Kruse, L. Experimental identification of specificity determinants in the domain linker of a LacI/GalR protein: bioinformatics-based predictions generate true positives and false negatives. Proteins 73, 941–957 (2008).
Taylor, N. D. et al. Engineering an allosteric transcription factor to respond to new ligands. Nat. Methods 13, 177–183 (2016).
Acknowledgements
We thank E. Cameron for his critical review and editing of the manuscript. The work was supported by the Wyss Institute for Biologically Inspired Engineering, the Paul G. Allen Frontiers Group, the Defense Threat Reduction Agency grant HDTRA1-14-1-0006, and Air Force Office of Scientific Research grant FA9550-14-1-0060. J.W.L. was also supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT and Future Planning (2018R1C1B3007409), and by the Marine Biotechnology Program (Marine BioMaterials Research Center) funded by the Ministry of Oceans and Fisheries, Korea. C.T.Y.C. was also supported by the University of Texas System Rising STARs Program and by the Welch Foundation (grant # BP-0037).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Lee, J.W., Chan, C.T.Y., Slomovic, S. et al. Next-generation biocontainment systems for engineered organisms. Nat Chem Biol 14, 530–537 (2018). https://doi.org/10.1038/s41589-018-0056-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-018-0056-x
This article is cited by
-
A bumpy road ahead for genetic biocontainment
Nature Communications (2024)
-
Synthetic microbiology in sustainability applications
Nature Reviews Microbiology (2024)
-
Advanced probiotics: bioengineering and their therapeutic application
Molecular Biology Reports (2024)
-
Engineering the gut microbiome
Nature Reviews Bioengineering (2023)
-
A robust yeast biocontainment system with two-layered regulation switch dependent on unnatural amino acid
Nature Communications (2023)