Abstract
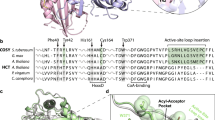
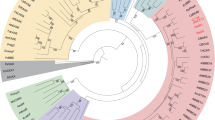
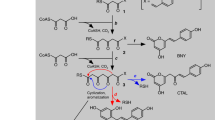
The emergence of catalysis in a noncatalytic protein scaffold is a rare, unexplored event. Chalcone isomerase (CHI), a key enzyme in plant flavonoid biosynthesis, is presumed to have evolved from a nonenzymatic ancestor related to the widely distributed fatty-acid binding proteins (FAPs) and a plant protein family with no isomerase activity (CHILs). Ancestral inference supported the evolution of CHI from a protein lacking isomerase activity. Further, we identified four alternative founder mutations, i.e., mutations that individually instated activity, including a mutation that is not phylogenetically traceable. Despite strong epistasis in other cases of protein evolution, CHI’s laboratory reconstructed mutational trajectory shows weak epistasis. Thus, enantioselective CHI activity could readily emerge despite a catalytically inactive starting point. Accordingly, X-ray crystallography, NMR, and molecular dynamics simulations reveal reshaping of the active site toward a productive substrate-binding mode and repositioning of the catalytic arginine that was inherited from the ancestral fatty-acid binding proteins.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Change history
14 May 2018
In the version of this article originally published, the number for the equal contributions footnote was missing for Miriam Kaltenbach and Jason R. Burke in the author list. The error has been corrected in the PDF and print versions of this article.
References
Adrain, C. & Freeman, M. New lives for old: evolution of pseudoenzyme function illustrated by iRhoms. Nat. Rev. Mol. Cell Biol. 13, 489–498 (2012).
Ortmayer, M. et al. An oxidative N-demethylase reveals PAS transition from ubiquitous sensor to enzyme. Nature 539, 593–597 (2016).
Taga, M. E., Larsen, N. A., Howard-Jones, A. R., Walsh, C. T. & Walker, G. C. BluB cannibalizes flavin to form the lower ligand of vitamin B12. Nature 446, 449–453 (2007).
Tam, R. & Saier, M. H. Jr. A bacterial periplasmic receptor homologue with catalytic activity: cyclohexadienyl dehydratase of Pseudomonas aeruginosa is homologous to receptors specific for polar amino acids. Res. Microbiol. 144, 165–169 (1993).
Yuhara, K., Yonehara, H., Hattori, T., Kobayashi, K. & Kirimura, K. Enzymatic characterization and gene identification of aconitate isomerase, an enzyme involved in assimilation of trans-aconitic acid, from Pseudomonas sp. WU-0701. FEBS J. 282, 4257–4267 (2015).
Koes, R. E., Quattrocchio, F. & Mol, J. N. M. The flavonoid biosynthetic pathway in plants: function and evolution. BioEssays 16, 123–132 (1994).
Ngaki, M. N. et al. Evolution of the chalcone-isomerase fold from fatty-acid binding to stereospecific catalysis. Nature 485, 530–533 (2012).
Morita, Y. et al. A chalcone isomerase-like protein enhances flavonoid production and flower pigmentation. Plant J. 78, 294–304 (2014).
Jiang, W. et al. Role of a chalcone isomerase-like protein in flavonoid biosynthesis in Arabidopsis t haliana. J. Exp. Bot. 66, 7165–7179 (2015).
Yang, Z., Kumar, S. & Nei, M. A new method of inference of ancestral nucleotide and amino acid sequences. Genetics 141, 1641–1650 (1995).
Jez, J. M., Bowman, M. E. & Noel, J. P. Role of hydrogen bonds in the reaction mechanism of chalcone isomerase. Biochemistry 41, 5168–5176 (2002).
Bar-Rogovsky, H. et al. Assessing the prediction fidelity of ancestral reconstruction by a library approach. Protein Eng. Des. Sel. 28, 507–518 (2015).
Eick, G. N., Bridgham, J. T., Anderson, D. P., Harms, M. J. & Thornton, J. W. Robustness of reconstructed ancestral protein functions to statistical uncertainty. Mol. Biol. Evol. 34, 247–261 (2017).
Randall, R. N., Radford, C. E., Roof, K. A., Natarajan, D. K. & Gaucher, E. A. An experimental phylogeny to benchmark ancestral sequence reconstruction. Nat. Commun. 7, 12847 (2016).
Tokuriki, N. et al. Diminishing returns and tradeoffs constrain the laboratory optimization of an enzyme. Nat. Commun. 3, 1257 (2012).
Breen, M. S., Kemena, C., Vlasov, P. K., Notredame, C. & Kondrashov, F. A. Epistasis as the primary factor in molecular evolution. Nature 490, 535–538 (2012).
de Visser, J. A., Cooper, T. F. & Elena, S. F. The causes of epistasis. Proc. Biol. Sci. 278, 3617–3624 (2011).
Harms, M. J. & Thornton, J. W. Evolutionary biochemistry: revealing the historical and physical causes of protein properties. Nat. Rev. Genet. 14, 559–571 (2013).
Kaltenbach, M. & Tokuriki, N. Dynamics and constraints of enzyme evolution. J. Exp. Zool. B Mol. Dev. Evol. 322, 468–487 (2014).
McCandlish, D. M., Rajon, E., Shal, P., Ding, Y. & Plotkin, J. B. The role of epistasis in protein evolution. Nature 497, E1–2; discussion E2–3 (2013).
Poelwijk, F. J., Kiviet, D. J., Weinreich, D. M. & Tans, S. J. Empirical fitness landscapes reveal accessible evolutionary paths. Nature 445, 383–386 (2007).
Whitlock, M. C., Phillips, P. C., Moore, F. B.-G. & Tonsor, S. J. Multiple fitness peaks and epistasis. Annu. Rev. Ecol. Syst. 26, 601–629 (1995).
Salverda, M. L. et al. Initial mutations direct alternative pathways of protein evolution. PLoS Genet. 7, e1001321 (2011).
Tokuriki, N., Stricher, F., Schymkowitz, J., Serrano, L. & Tawfik, D. S. The stability effects of protein mutations appear to be universally distributed. J. Mol. Biol. 369, 1318–1332 (2007).
Kaltenbach, M., Jackson, C. J., Campbell, E. C., Hollfelder, F. & Tokuriki, N. Reverse evolution leads to genotypic incompatibility despite functional and active site convergence. eLife 4, e06492 (2015).
Dickinson, B. C., Leconte, A. M., Allen, B., Esvelt, K. M. & Liu, D. R. Experimental interrogation of the path dependence and stochasticity of protein evolution using phage-assisted continuous evolution. Proc. Natl Acad. Sci. USA 110, 9007–9012 (2013).
Jez, J. M., Bowman, M. E., Dixon, R. A. & Noel, J. P. Structure and mechanism of the evolutionarily unique plant enzyme chalcone isomerase. Nat. Struct. Biol. 7, 786–791 (2000).
Farrow, N. A. et al. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry 33, 5984–6003 (1994).
Thomsen, M. et al. Structure and catalytic mechanism of the evolutionarily unique bacterial chalcone isomerase. Acta Crystallogr. D Biol. Crystallogr. 71, 907–917 (2015).
Gumulya, Y. & Gillam, E. M. Exploring the past and the future of protein evolution with ancestral sequence reconstruction: the ‘retro’ approach to protein engineering. Biochem. J. 474, 1–19 (2017).
Weng, J. K. & Chapple, C. The origin and evolution of lignin biosynthesis. New Phytol. 187, 273–285 (2010).
Bar-Even, A. & Salah Tawfik, D. Engineering specialized metabolic pathways-is there a room for enzyme improvements? Curr. Opin. Biotechnol. 24, 310–319 (2013).
Keller, M. A., Piedrafita, G. & Ralser, M. The widespread role of non-enzymatic reactions in cellular metabolism. Curr. Opin. Biotechnol. 34, 153–161 (2015).
Trudeau, D. L., Kaltenbach, M. & Tawfik, D. S. On the potential origins of the high stability of reconstructed ancestral proteins. Mol. Biol. Evol. 33, 2633–2641 (2016).
Noor, S. et al. Intramolecular epistasis and the evolution of a new enzymatic function. PLoS One 7, e39822 (2012).
Lozovsky, E. R. et al. Stepwise acquisition of pyrimethamine resistance in the malaria parasite. Proc. Natl. Acad. Sci. USA 106, 12025–12030 (2009).
Kiss, G., Çelebi-Ölçüm, N., Moretti, R., Baker, D. & Houk, K. N. Computational enzyme design. Angew. Chem. Int. Edn. Engl. 52, 5700–5725 (2013).
Kries, H., Blomberg, R. & Hilvert, D. De novo enzymes by computational design. Curr. Opin. Chem. Biol. 17, 221–228 (2013).
Lassila, J. K. Conformational diversity and computational enzyme design. Curr. Opin. Chem. Biol. 14, 676–682 (2010).
Blomberg, R. et al. Precision is essential for efficient catalysis in an evolved Kemp eliminase. Nature 503, 418–421 (2013).
Khersonsky, O. et al. Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59. Proc. Natl Acad. Sci. USA 109, 10358–10363 (2012).
Matasci, N. et al. Data access for the 1,000 Plants (1KP) project. Gigascience 3, 17 (2014).
Armougom, F. et al. Expresso: automatic incorporation of structural information in multiple sequence alignments using 3D-Coffee. Nucleic Acids Res. 34, W604–W608 (2006).
Ronquist, F. & Huelsenbeck, J. P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 (2003).
Cronk, Q. C. Plant evolution and development in a post-genomic context. Nat. Rev. Genet. 2, 607–619 (2001).
Ashkenazy, H. et al. FastML: a web server for probabilistic reconstruction of ancestral sequences. Nucleic Acids Res. 40, W580–W584 (2012).
Miranda, C. L. et al. Antioxidant and prooxidant actions of prenylated and nonprenylated chalcones and flavanones in vitro. J. Agric. Food Chem. 48, 3876–3884 (2000).
Ashkenazy, H. et al. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 38, W529–W533 (2010).
Herman, A. & Tawfik, D. S. Incorporating synthetic oligonucleotides via gene reassembly (ISOR): a versatile tool for generating targeted libraries. Protein Eng. Des. Sel. 20, 219–226 (2007).
Zhao, H., Giver, L., Shao, Z., Affholter, J. A. & Arnold, F. H. Molecular evolution by staggered extension process (StEP) in vitro recombination. Nat. Biotechnol. 16, 258–261 (1998).
Battye, T. G. et al. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 67, 271–281 (2011).
Evans, P. Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 (2006).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
van den Bedem, H., Dhanik, A., Latombe, J. C. & Deacon, A. M. Modeling discrete heterogeneity in X-ray diffraction data by fitting multi-conformers. Acta Crystallogr. D Biol. Crystallogr. 65, 1107–1117 (2009).
Goddard, T. D. & Kneller, D. G. Sparky 3. (University of California, San Francisco, 2008).
Bieri, M., d’Auvergne, E. J. & Gooley, P. R. relaxGUI: a new software for fast and simple NMR relaxation data analysis and calculation of ps-ns and µs motion of proteins. J. Biomol. NMR 50, 147–155 (2011).
Delaglio, F. et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995).
Fiser, A. & Sali, A. Modeller: generation and refinement of homology-based protein structure models. Methods Enzymol. 374, 461–491 (2003).
Sondergaard, C. R., Olsson, M. H., Rostkowski, M. & Jensen, J. H. Improved treatment of ligands and coupling effects in empirical calculation and rationalization of pK a values. J. Chem. Theory Comput. 7, 2284–2295 (2011).
Case, D. A. et al. AMBER 2016. (University of California, San Francisco, 2016).
Maier, J. A. et al. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 11, 3696–3713 (2015).
Wang, J. et al. Development and testing of a general amber force field. J. Comput. Chem. 25, 1157–1174 (2004).
Frisch, M. J. et al. Gaussian 09, Revision D.01. (Gaussian, Inc., Wallingford, CT, 2016).
Cieplak, P., Cornell, W. D., Bayly, C. & Kollman, P. A. Application of the multimolecule and multiconformational RESP methodology to biopolymers: charge derivation for DNA, RNA, and proteins. J. Comput. Chem. 16, 1357–1377 (1995).
Jorgensen, W. L. et al. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983).
Berendsen, H. J. C. et al. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81, 3684–3690 (1984).
Ryckaert, J.-P., Ciccotti, G. & Berendsen, H. J. C. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 23, 327–341 (1977).
Feller, S. E., Zhang, Y., Pastor, R. W. & Brooks, B. R. Constant pressure molecular dynamics simulation: the Langevin piston method. J. Chem. Phys. 103, 4613–4621 (1995).
Darden, T., York, D. & Pederson, L. Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092 (1993).
Humphrey, W., Dalke, A. & Schulten, K. VMD: visual molecular dynamics. J. Mol. Graph. 14, 27–38 (1996).
McGibbon, R. T. et al. MDTraj: a modern open library for the analysis of molecular dynamics trajectories. Biophys. J. 109, 1528–1532 (2015).
Pettersen, E. F. et al. UCSF Chimera-a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Acknowledgements
We thank K.-P. Cherukuri for help with the synthesis of chalconaringenin, B. Duggan and X. Huang for assistance with NMR, G. Louie for assistance with protein X-ray data collection and processing, and G. Cortina for help with analyzing the simulations. This work was funded by the Israel Science Foundation Grant 980/14 and the Sasson & Marjorie Peress Philanthropic Fund (D.S.T.); the United States National Science Foundation grant EEC-0813570 (J.P.N.); the Knut and Alice Wallenberg Foundation, Wenner-Gren Foundations and the European Research Council (S.C.L.K.). Computer time was provided by the Swedish National Infrastructure for Computing. J.P.N. is the Arthur and Julie Woodrow Chair and a Howard Hughes Medical Institute investigator. D.S.T. is the Nella and Leon Benoziyo Professor of Biochemistry.
Author information
Authors and Affiliations
Contributions
M.K. performed ancestral inference with assistance from A.R. M.K. performed directed evolution. M.K., M.D., and J.R.B. performed mutagenesis, protein expression, stable isotope labeling, and biochemical characterization of the proteins. J.R.B., M.K., D.S.T. and J.P.N. performed and analyzed protein X-ray crystallography and NMR. A.P. and S.C.L.K. performed and analyzed MD simulations with assistance from F.S.M. D.S.T. and J.P.N. planned and directed the project, and, together with M.K., J.R.B., A.P., and S.C.L.K., designed the experiments. M.K., J.R.B., J.P.N. and D.S.T. wrote and edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1–12, Supplementary Figures 1– 20 and Supplementary Notes 1– 3
Supplementary Dataset 1
>AUGV-6826_P_4dokB
Supplementary Dataset 2
Posterior probabilities P(amino acid) in ancestral reconstruction including indels.
Rights and permissions
About this article
Cite this article
Kaltenbach, M., Burke, J.R., Dindo, M. et al. Evolution of chalcone isomerase from a noncatalytic ancestor. Nat Chem Biol 14, 548–555 (2018). https://doi.org/10.1038/s41589-018-0042-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-018-0042-3
This article is cited by
-
Genome-wide identification of rubber tree pathogenesis-related 10 (PR-10) proteins with biological relevance to plant defense
Scientific Reports (2024)
-
The evolutionary origin of naturally occurring intermolecular Diels-Alderases from Morus alba
Nature Communications (2024)
-
Extant Sequence Reconstruction: The Accuracy of Ancestral Sequence Reconstructions Evaluated by Extant Sequence Cross-Validation
Journal of Molecular Evolution (2024)
-
Loop dynamics and the evolution of enzyme activity
Nature Reviews Chemistry (2023)
-
Emergence of a proton exchange-based isomerization and lactonization mechanism in the plant coumarin synthase COSY
Nature Communications (2023)