Abstract
Homotypic chromatin interactions and loop extrusion are thought to be the two main drivers of mammalian chromosome folding. Here we tested the role of RNA polymerase II (RNAPII) across different scales of interphase chromatin organization in a cellular system allowing for its rapid, auxin-mediated degradation. We combined Micro-C and computational modeling to characterize subsets of loops differentially gained or lost upon RNAPII depletion. Gained loops, extrusion of which was antagonized by RNAPII, almost invariably formed by engaging new or rewired CTCF anchors. Lost loops selectively affected contacts between enhancers and promoters anchored by RNAPII, explaining the repression of most genes. Surprisingly, promoter–promoter interactions remained essentially unaffected by polymerase depletion, and cohesin occupancy was sustained. Together, our findings reconcile the role of RNAPII in transcription with its direct involvement in setting-up regulatory three-dimensional chromatin contacts genome wide, while also revealing an impact on cohesin loop extrusion.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
NGS data generated in this study are available via the NCBI Gene Expression Omnibus repository under accession number GSE178593 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE178593). All other data used for analyses come from our previous study39, and are available under accession number GSE160321 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE160321). Source data are provided with this paper.
Code availability
All custom code used for Micro-C analysis is available at https://github.com/shuzhangcourage/Micro-C-CUT-tag/tree/v1.0.0 (ref. 73), and all the code used for our Molecular Dynamics simulations is available at https://zenodo.org/record/7674875#.Y_n5zXbMLBR (ref. 74).
References
Aboelnour, E. & Bonev, B. Decoding the organization, dynamics, and function of the 4D genome. Dev. Cell 56, 1562–1573 (2021).
Razin, S. V. & Kantidze, O. L. The twisted path of the 3D genome: where does it lead? Trends Biochem. Sci. 47, 736–744 (2022).
Xiang, J. F. & Corces, V. G. Regulation of 3D chromatin organization by CTCF. Curr. Opin. Genet. Dev. 67, 33–40 (2021).
van Ruiten, M. S. & Rowland, B. D. On the choreography of genome folding: a grand pas de deux of cohesin and CTCF. Curr. Opin. Cell Biol. 70, 84–90 (2021).
Beagan, J. A. & Phillips-Cremins, J. E. On the existence and functionality of topologically associating domains. Nat. Genet. 52, 8–16 (2020).
Rao, S. S. P. et al. Cohesin loss eliminates all loop domains. Cell 17, 305–320 (2017).
Fudenberg, G. et al. Formation of chromosomal domains by loop extrusion. Cell Rep. 15, 2038–2049 (2016).
Davidson, I. F. et al. DNA loop extrusion by human cohesin. Science 366, 1338–1345 (2019).
Kim, Y., Shi, Z., Zhang, H., Finkelstein, I. J. & Yu, H. Human cohesin compacts DNA by loop extrusion. Science 366, 1345–1349 (2019).
Nora, E. P. et al. Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell 169, 930–944 (2017).
Wutz, G. et al. Topologically associating domains and chromatin loops depend on cohesin and are regulated by CTCF, WAPL, and PDS5 proteins. EMBO J. 36, 3573–3599 (2017).
Wutz, G. et al. ESCO1 and CTCF enable formation of long chromatin loops by protecting cohesinSTAG1 from WAPL. eLife 9, e52091 (2020).
Li, Y. et al. The structural basis for cohesin-CTCF-anchored loops. Nature 578, 472–476 (2020).
Rao, S. S. et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014).
Schwarzer, W. et al. Two independent modes of chromatin organization revealed by cohesin removal. Nature 551, 51–56 (2017).
Haarhuis, J. H. I. et al. The cohesin release factor WAPL restricts chromatin loop extension. Cell 169, 693–707 (2017).
Gabriele, M. et al. Dynamics of CTCF- and cohesin-mediated chromatin looping revealed by live-cell imaging. Science 376, 496–501 (2022).
Nuebler, J., Fudenberg, G., Imakaev, M., Abdennur, N. & Mirny, L. A. Chromatin organization by an interplay of loop extrusion and compartmental segregation. Proc. Natl Acad. Sci. USA 115, E6697–E6706 (2018).
Rada-Iglesias, A., Grosveld, F. G. & Papantonis, A. Forces driving the three-dimensional folding of eukaryotic genomes. Mol. Syst. Biol. 14, e8214 (2018).
Papantonis, A. & Cook, P. R. Fixing the model for transcription: the DNA moves, not the polymerase. Transcription 2, 41–44 (2011).
Lee, K. & Blobel, G. A. Chromatin architecture underpinning transcription elongation. Nucleus 7, 1–8 (2016).
Cook, P. R. & Marenduzzo, D. Transcription-driven genome organization: a model for chromosome structure and the regulation of gene expression tested through simulations. Nucleic Acids Res. 46, 9895–9906 (2018).
Racko, D., Benedetti, F., Dorier, J. & Stasiak, A. Transcription-induced supercoiling as the driving force of chromatin loop extrusion during formation of TADs in interphase chromosomes. Nucleic Acids Res. 46, 1648–1660 (2018).
Mitchell, J. A. & Fraser, P. Transcription factories are nuclear subcompartments that remain in the absence of transcription. Genes Dev. 22, 20–25 (2008).
Palstra, R. J. et al. Maintenance of long-range DNA interactions after inhibition of ongoing RNA polymerase II transcription. PLoS ONE 3, e1661 (2008).
Ke, Y. et al. 3D chromatin structures of mature gametes and structural reprogramming during mammalian embryogenesis. Cell 170, 367–381 (2017).
Barutcu, A. R., Blencowe, B. J. & Rinn, J. L. Differential contribution of steady-state RNA and active transcription in chromatin organization. EMBO Rep. 20, e48068 (2019).
Brant, L. et al. Exploiting native forces to capture chromosome conformation in mammalian cell nuclei. Mol. Syst. Biol. 12, 891 (2016).
El Khattabi, L. et al. A pliable Mediator acts as a functional rather than an architectural bridge between promoters and enhancers. Cell 178, 1145–1158 (2019).
Jiang, Y. et al. Genome-wide analyses of chromatin interactions after the loss of Pol I, Pol II, and Pol III. Genome Biol. 21, 158 (2020).
Sun, F. et al. The Pol II preinitiation complex (PIC) influences Mediator binding but not promoter-enhancer looping. Genes Dev. 35, 1175–1189 (2021).
Haarhuis, J. H. I. et al. A Mediator-cohesin axis controls heterochromatin domain formation. Nat. Commun. 13, 754 (2022).
Hsieh, T. S. et al. Resolving the 3D landscape of transcription-linked mammalian chromatin folding. Mol. Cell 78, 539–553 (2020).
Krietenstein, N. et al. Ultrastructural details of mammalian chromosome architecture. Mol. Cell 78, 554–565 (2020).
Hua, P. et al. Defining genome architecture at base-pair resolution. Nature 595, 125–129 (2021).
Cramer, P. Organization and regulation of gene transcription. Nature 573, 45–54 (2019).
Ramasamy, S. et al. The mediator complex regulates enhancer–promoter interactions. Preprint at bioRxiv, https://doi.org/10.1101/2022.06.15.496245 (2022).
Nagashima, R. et al. Single nucleosome imaging reveals loose genome chromatin networks via active RNA polymerase II. J. Cell Biol. 218, 1511–1530 (2019).
Zhang, S. et al. RNA polymerase II is required for spatial chromatin reorganization following exit from mitosis. Sci. Adv. 7, eabg8205 (2021).
Buckle, A., Brackley, C. A., Boyle, S., Marenduzzo, D. & Gilbert, N. Polymer simulations of heteromorphic chromatin predict the 3D folding of complex genomic loci. Mol. Cell 72, 786–797 (2018).
Fiorillo, L. et al. Inference of chromosome 3D structures from GAM data by a physics computational approach. Methods 181–182, 70–79 (2020).
Busslinger, G. A. et al. Cohesin is positioned in mammalian genomes by transcription, CTCF and Wapl. Nature 544, 503–507 (2017).
Zhu, Y., Denholtz, M., Lu, H. & Murre, C. Calcium signaling instructs NIPBL recruitment at active enhancers and promoters via distinct mechanisms to reconstruct genome compartmentalization. Genes Dev. 35, 65–81 (2021).
Rinaldi, L. et al. The glucocorticoid receptor associates with the cohesin loader NIPBL to promote long-range gene regulation. Sci. Adv. 8, eabj8360 (2022).
Rinzema, N. J. et al. Building regulatory landscapes reveals that an enhancer can recruit cohesin to create contact domains, engage CTCF sites and activate distant genes. Nat. Struct. Mol. Biol. 29, 563–574 (2022).
Heinz, S. et al. Transcription elongation can affect genome 3D structure. Cell 174, 1522–1536 (2018).
Olan, I. et al. Transcription-dependent cohesin repositioning rewires chromatin loops in cellular senescence. Nat. Commun. 11, 6049 (2020).
Valton, A. L. et al. A cohesin traffic pattern genetically linked to gene regulation. Nat. Struct. Mol. Biol. 29, 1239–1251 (2022).
Banigan, E. J. et al. Transcription shapes 3D chromatin organization by interacting with loop extrusion. Proc. Natl Acad. Sci. USA 120, e2210480120 (2023).
Rosencrance, C. D. et al. Chromatin hyperacetylation impacts chromosome folding by forming a nuclear subcompartment. Mol. Cell 78, 112–126 (2020).
Ferrai, C. et al. RNA polymerase II primes Polycomb-repressed developmental genes throughout terminal neuronal differentiation. Mol. Syst. Biol. 13, 946 (2017).
Rhodes, J. D. P. et al. Cohesin disrupts polycomb-dependent chromosome interactions in embryonic stem cells. Cell Rep. 30, 820–835 (2020).
Penagos-Puig, A. et al. RNA polymerase II pausing contributes to maintain chromatin organization in erythrocytes. Preprint at bioRxiv https://doi.org/10.1101/2022.06.16.496295 (2022).
Andersson, R., Sandelin, A. & Danko, C. G. A unified architecture of transcriptional regulatory elements. Trends Genet. 31, 426–433 (2015).
Casa, V. et al. Redundant and specific roles of cohesin STAG subunits in chromatin looping and transcriptional control. Genome Res. 30, 515–527 (2020).
Liu, N. Q. et al. WAPL maintains a cohesin loading cycle to preserve cell-type-specific distal gene regulation. Nat. Genet. 53, 100–109 (2021).
Hakimi, M. A. et al. A chromatin remodelling complex that loads cohesin onto human chromosomes. Nature 418, 994–998 (2002).
Kagey, M. H. et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467, 430–435 (2010).
Mattingly, M. et al. Mediator recruits the cohesin loader Scc2 to RNA Pol II-transcribed genes and promotes sister chromatid cohesion. Curr. Biol. 32, 2884–2896 (2022).
Liu, Y. et al. Systematic inference and comparison of multi-scale chromatin sub-compartments connects spatial organization to cell phenotypes. Nat. Commun. 12, 2439 (2021).
Greenwald, W. W. et al. Pgltools: a genomic arithmetic tool suite for manipulation of Hi-C peak and other chromatin interaction data. BMC Bioinformatics 18, 207 (2017).
Yu, G., Wang, L. G. & He, Q. Y. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31, 2382–2383 (2015).
Whyte, W. A. et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319 (2013).
Flyamer, I. M., Illingworth, R. S. & Bickmore, W. A. Coolpup.py: versatile pile-up analysis of Hi-C data. Bioinformatics 36, 2980–2985 (2020).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Grant, C. E., Bailey, T. L. & Noble, W. S. FIMO: scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018 (2011).
Ramírez, F., Dündar, F., Diehl, S., Grüning, B. A. & Manke, T. DeepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 42, W187–W191 (2014).
Watrin, E. et al. Human Scc4 is required for cohesin binding to chromatin, sister-chromatid cohesion, and mitotic progression. Curr. Biol. 16, 863–874 (2006).
Reynwar, B. J. et al. Aggregation and vesiculation of membrane proteins by curvature-mediated interactions. Nature 447, 461–464 (2007).
Chiariello, A. M., Annunziatella, C., Bianco, S., Esposito, A. & Nicodemi, M. Polymer physics of chromosome large-scale 3D organisation. Sci. Rep. 6, 29775 (2016).
Barbieri, M. et al. Active and poised promoter states drive folding of the extended HoxB locus in mouse embryonic stem cells. Nat. Struct. Mol. Biol. 24, 515–524 (2017).
Yang, T. et al. HiCRep: assessing the reproducibility of Hi-C data using a stratum-adjusted correlation coefficient. Genome Res. 27, 1939–1949 (2017).
Zhang, S. Code used for Micro-C and CUT&Tag data analysis. Zenodo https://doi.org/10.5281/zenodo.7656606 (2023).
Barbieri, M. Code used for MD simulations. Zenodo https://doi.org/10.5281/zenodo.7674875 (2023).
Acknowledgements
We thank M. Oudelaar, K. Wendt and all members of the Papantonis lab for discussions, and the Maeshima lab (NIG, Japan) for the DLD-1 mAID-RPB1 cells. This work was funded by the Deutsche Forschungsgemeinschaft (DFG) via the SPP2202 (PA 2456/11-2) and SPP2191 Priority Programs (PA 2456/17-1), and the TRR81 TransRegio program (INST 160/697-1), all awarded to A.P. S.Z. is supported by a China Scholarship Council fellowship. S.Z. and N.Ü. are members of the International Max Planck Research School for Genome Science. The funders had no role in the study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
S.Z. performed all bioinformatics analyses; N.Ü. performed all experiments; M.B. performed the computational modeling; and A.P. conceived and supervised the study and compiled the manuscript with input from all co-authors.
Corresponding author
Ethics declarations
Competing interests
All authors declare no competing interest.
Peer review
Peer review information
Nature Genetics thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
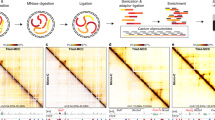
Extended Data Fig. 1 Effects of RNAPII depletion on chromatin organization and protein levels.
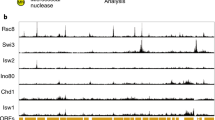
a, Left: Schematic of the biallelic tagging strategy in the endogenous POLR2A loci. Right: Fractionation blots showing the levels of RPB1 and Ser5-phosphorylated RNAPII, Mediator subunit 24, and Lamin B1 from DLD1-mAID-RBP1 cells treated or not with auxin to deplete RNAPII. HSC70 levels provide a control. Blots have been replicated at least twice. b, Representative tracks of CUT&Tag signal for H3K27me3, H3K27ac, SMC1A, and CTCF from control (left) and auxin-treated DLD1-mAID-RBP1 cells (right) along 0.55 Mbp of chr1. c, Heatmaps of nucleosome occupancy deduced from Micro-C data, of chromatin accessibility deduced from ATAC-seq, and of CTCF and SMC1A occupancy deduced from CUT&Tag around CTCF loop anchors before (ctrl) and after RNAPII degradation (+auxin). d, As in panel c, but showing scaled ATAC-seq signal around gene promoters and enhancers. e, As in panel a, but for Ser5-phosphorylated RNAPII, NIPBL, SMC1A, CTCF, and H3K27me3 levels in the soluble and chromatin fractions of DLD-1 cells. HSC70 levels provide a control. Blots have been replicated at least twice. f, Boxplots depicting the distribution of genes containing (genes with +aux loops) or not (ctrl genes) gained loop anchors upon RNAPII depletion. In the plots, center lines represent the median value, box-limits the 25th and 75th percentiles, and whiskers extend 1.5x each box’s interquartile range. *P < 0.01, two-sided Wilcoxon-Mann-Whitney test.
Extended Data Fig. 2 Effects of RNAPII depletion on the 3D organization of facultative heterochromatin.
a, Micro-C contact maps from control (left) and auxin-treated cells (right) in two exemplary genomic regions of chr1 at 2-kbp resolution aligned to H3K27me3, H3K27ac, CTCF, and SMC1A CUT&Tag signal tracks. Loops called for each region and condition are also shown by spider plots (bottom). b, Aggregate plots of all H3K27me3-anchored loops emerging in auxin-treated cells. c, Bar plot showing per cent of gained loops with one or two H3K27me3 anchors or with H3K27me3 in the next-door genomic bin (that is, within <10 kbp from the anchor).
Extended Data Fig. 3 Changes in loops and stripes following RNAPII depletion.
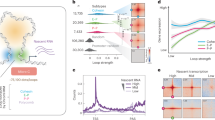
a, Micro-C contact maps from control (left) and auxin-treated cells (right) in an exemplary genomic region on chr1 at 4-kbp resolution aligned to H3K27ac, CTCF, and SMC1A CUT&Tag signal tracks. Loops called for each region and condition are also shown by spider plots (bottom). b, Plots of interaction frequency decay as a function of genomic distance from control and auxin-treated cells (top) and their first derivative (bottom). c, Aggregate plots of gene promoter-promoter (P-P) or enhancer- promoter loops (E-P) in control and auxin-treated cells that involve (+CTCF) or not CTCF (wo CTCF) in at least one anchor. d, Line plot showing mean SMC1A CUT&Tag signal from control and auxin-treated cells in the 6 kbp around H3K27ac peaks from 590 super-enhancers. e, As in panel c, but for H3K27ac signal around active gene promoters or enhancers. f, Average plots showing mean signal of stripes with one CTCF and one transcriptional anchor before (ctrl) and after RNAPII depletion (+auxin). Zoom-in: Aggregate plots for loops at the end of the stripes. g, As in panel c, but for shared loops that rewire one anchor by <20 kbp (see cartoon). h, Boxplots depicting the lengths of genes downregulated upon RNAPII depletion that are linked (genes with lost E-P loops) or not (all other genes) to lost E-P loops. In the plots, center lines represent the medians, box-limits the 25th and 75th percentiles, and whiskers extend 1.5x each box’s interquartile range. *: P < 0.01, two-sided Wilcoxon-Mann-Whitney test.
Supplementary information
Supplementary Information
Supplementary Figs. 1 and 2 and Supplementary Tables 1 and 2.
Source data
Source Data Extended Data Fig. 1
Unprocessed/uncropped western blots from Extended Data Fig. 1a,e.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, S., Übelmesser, N., Barbieri, M. et al. Enhancer–promoter contact formation requires RNAPII and antagonizes loop extrusion. Nat Genet 55, 832–840 (2023). https://doi.org/10.1038/s41588-023-01364-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-023-01364-4
This article is cited by
-
Transcription regulation by long non-coding RNAs: mechanisms and disease relevance
Nature Reviews Molecular Cell Biology (2024)
-
H4K16ac activates the transcription of transposable elements and contributes to their cis-regulatory function
Nature Structural & Molecular Biology (2023)
-
RNA Pol II enters the ring of cohesin-mediated loop extrusion
Nature Genetics (2023)
-
The Mediator complex regulates enhancer-promoter interactions
Nature Structural & Molecular Biology (2023)
-
Region Capture Micro-C reveals coalescence of enhancers and promoters into nested microcompartments
Nature Genetics (2023)