Abstract
Endometriosis is a common condition associated with debilitating pelvic pain and infertility. A genome-wide association study meta-analysis, including 60,674 cases and 701,926 controls of European and East Asian descent, identified 42 genome-wide significant loci comprising 49 distinct association signals. Effect sizes were largest for stage 3/4 disease, driven by ovarian endometriosis. Identified signals explained up to 5.01% of disease variance and regulated expression or methylation of genes in endometrium and blood, many of which were associated with pain perception/maintenance (SRP14/BMF, GDAP1, MLLT10, BSN and NGF). We observed significant genetic correlations between endometriosis and 11 pain conditions, including migraine, back and multisite chronic pain (MCP), as well as inflammatory conditions, including asthma and osteoarthritis. Multitrait genetic analyses identified substantial sharing of variants associated with endometriosis and MCP/migraine. Targeted investigations of genetically regulated mechanisms shared between endometriosis and other pain conditions are needed to aid the development of new treatments and facilitate early symptomatic intervention.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Summary results for the top 10,000 SNPs included in the entire Genome-wide Association Study (GWAS) meta-analysis are provided in Supplementary Table 33. Summary statistics from the endometriosis meta-analysis excluding 23andMe are available from EBI GWAS Catalog Study Accession GCST90205183. GWAS summary statistics from 23andMe, Inc. were made available under a data use agreement that protects participant privacy. Please contact dataset-request@23andme.com or visit research.23andMe.com/collaborate for more information and to apply to access the data. The UK endometrium eQTL dataset (n = 163) 1 MB around each 42 lead SNPs is provided in Supplementary Tables 34 and 35.
Code availability
We utilized publicly available software in all the analyses. These are listed with appropriate citations in the methods.
References
Zondervan, K. T., Becker, C. M. & Missmer, S. A. Endometriosis. N. Engl. J. Med. 382, 1244–1256 (2020).
Nnoaham, K. E. et al. Impact of endometriosis on quality of life and work productivity: a multicenter study across ten countries. Fertil. Steril. 96, 366–373 (2011).
Revised, A. S. R. M. American society for reproductive medicine classification of endometriosis: 1996. Fertil. Steril. 67, 817–821 (1997).
Saha, R. et al. Heritability of endometriosis. Fertil. Steril. 104, 947–952 (2015).
Treloar, S. A., O’Connor, D. T., O’Connor, V. M. & Martin, N. G. Genetic influences on endometriosis in an Australian twin sample. sueT@qimr.edu.au. Fertil. Steril. 71, 701–710 (1999).
Lee, S. H. et al. Estimation and partitioning of polygenic variation captured by common SNPs for Alzheimer’s disease, multiple sclerosis and endometriosis. Hum. Mol. Genet. 22, 832–841 (2013).
Zondervan, K. T. et al. Endometriosis. Nat. Rev. Dis. Primers 4, 9 (2018).
Sapkota, Y. et al. Meta-analysis identifies five novel loci associated with endometriosis highlighting key genes involved in hormone metabolism. Nat. Commun. 8, 15539 (2017).
Painter, J. N. et al. Genome-wide association study identifies a locus at 7p15.2 associated with endometriosis. Nat. Genet. 43, 51–54 (2011).
Fung, J. N. et al. Genetic regulation of disease risk and endometrial gene expression highlights potential target genes for endometriosis and polycystic ovarian syndrome. Sci. Rep. 8, 11424 (2018).
Mortlock, S. et al. Tissue specific regulation of transcription in endometrium and association with disease. Hum. Reprod. 35, 377–393 (2020).
Võsa, U. et al. Unraveling the polygenic architecture of complex traits using blood eQTL meta-analysis. Nat. Genet. 53, 1300–1310 (2021).
McRae, A. F. et al. Identification of 55,000 replicated DNA methylation QTL. Sci. Rep. 8, 17605 (2018).
Mortlock, S. et al. Genetic regulation of methylation in human endometrium and blood and gene targets for reproductive diseases. Clin. Epigenetics 11, 49 (2019).
Zhu, Z. et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet. 48, 481–487 (2016).
Fung, J. N. et al. Functional evaluation of genetic variants associated with endometriosis near GREB1. Hum. Reprod. 30, 1263–1275 (2015).
Jones, A. V. et al. Genome-wide association analysis of pain severity in dysmenorrhea identifies association at chromosome 1p13.2, near the nerve growth factor locus. Pain 157, 2571–2581 (2016).
Barneo-Munoz, M. et al. Lack of GDAP1 induces neuronal calcium and mitochondrial defects in a knockout mouse model of charcot-marie-tooth neuropathy. PLoS Genet. 11, e1005115 (2015).
Ruth, K. S. et al. Using human genetics to understand the disease impacts of testosterone in men and women. Nat. Med. 26, 252–258 (2020).
Zhai, G. et al. Eight common genetic variants associated with serum DHEAS levels suggest a key role in ageing mechanisms. PLoS Genet. 7, e1002025 (2011).
Rahmani, A., Shoae-Hassani, A., Keyhanvar, P., Kheradmand, D. & Darbandi-Azar, A. Dehydroepiandrosterone stimulates nerve growth factor and brain derived neurotrophic factor in cortical neurons. Adv. Pharm. Sci. 2013, 506191 (2013).
Maninger, N., Wolkowitz, O. M., Reus, V. I., Epel, E. S. & Mellon, S. H. Neurobiological and neuropsychiatric effects of dehydroepiandrosterone (DHEA) and DHEA sulfate (DHEAS). Front. Neuroendocrinol. 30, 65–91 (2009).
Obata, K. & Noguchi, K. BDNF in sensory neurons and chronic pain. Neurosci. Res. 55, 1–10 (2006).
Browne, A. S. et al. Proteomic identification of neurotrophins in the eutopic endometrium of women with endometriosis. Fertil. Steril. 98, 713–719 (2012).
Wang, S. et al. BDNF and TrKB expression levels in patients with endometriosis and their associations with dysmenorrhoea. J. Ovarian Res. 15, 35 (2022).
Peng, B., Alotaibi, F. T., Sediqi, S., Bedaiwy, M. A. & Yong, P. J. Role of interleukin-1beta in nerve growth factor expression, neurogenesis and deep dyspareunia in endometriosis. Hum. Reprod. 35, 901–912 (2020).
Vitonis, A. F. et al. World endometriosis research foundation endometriosis phenome and biobanking harmonization project: II. Clinical and covariate phenotype data collection in endometriosis research. Fertil. Steril. 102, 1223–1232 (2014).
Nathan, A. et al. The Wilms tumor protein Wt1 contributes to female fertility by regulating oviductal proteostasis. Hum. Mol. Genet. 26, 1694–1705 (2017).
O’Mara, T. A. et al. Comprehensive genetic assessment of the ESR1 locus identifies a risk region for endometrial cancer. Endocr. Relat. Cancer 22, 851–861 (2015).
Marla, S. et al. Genetic risk factors for endometriosis near estrogen receptor 1 and coexpression of genes in this region in endometrium. Mol. Hum. Reprod. 27, gaaa082 (2021).
Smith, S. B. et al. Epistasis between polymorphisms in COMT, ESR1, and GCH1 influences COMT enzyme activity and pain. Pain 155, 2390–2399 (2014).
Martin, V. T. Ovarian hormones and pain response: a review of clinical and basic science studies. Gend. Med. 6, 168–192 (2009).
Smith, Y. R. et al. Pronociceptive and antinociceptive effects of estradiol through endogenous opioid neurotransmission in women. J. Neurosci. 26, 5777–5785 (2006).
Bulik-Sullivan, B. et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 47, 1236–1241 (2015).
Bulik-Sullivan, B. K. et al. LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47, 291–295 (2015).
Shafrir, A. L. et al. Risk for and consequences of endometriosis: a critical epidemiologic review. Best Pract. Res. Clin. Obstet. Gynaecol. 51, 1–15 (2018).
Missmer, S. A. et al. Reproductive history and endometriosis among premenopausal women. Obstet. Gynecol. 104, 965–974 (2004).
Sampson, J. A. Metastatic or embolic endometriosis, due to the menstrual dissemination of endometrial tissue into the venous circulation. Am. J. Pathol. 3, 93–110 (1927).
Barban, N. et al. Genome-wide analysis identifies 12 loci influencing human reproductive behavior. Nat. Genet. 48, 1462–1472 (2016).
As-Sanie, S. et al. Changes in regional gray matter volume in women with chronic pelvic pain: a voxel-based morphometry study. Pain 153, 1006–1014 (2012).
Coxon, L., Horne, A. W. & Vincent, K. Pathophysiology of endometriosis-associated pain: a review of pelvic and central nervous system mechanisms. Best Pract. Res. Clin. Obstet. Gynaecol. 51, 53–67 (2018).
Bajaj, P., Bajaj, P., Madsen, H. & Arendt-Nielsen, L. Endometriosis is associated with central sensitization: a psychophysical controlled study. J. Pain 4, 372–380 (2003).
Berkley, K. J., Rapkin, A. J. & Papka, R. E. The pains of endometriosis. Science 308, 1587–1589 (2005).
Vincent, K. et al. Dysmenorrhoea is associated with central changes in otherwise healthy women. Pain 152, 1966–1975 (2011).
Elliott, L. T. et al. Genome-wide association studies of brain imaging phenotypes in UK Biobank. Nature 562, 210–216 (2018).
Kamat, M. A. et al. PhenoScanner V2: an expanded tool for searching human genotype-phenotype associations. Bioinformatics 35, 4851–4853 (2019).
Turley, P. et al. Multi-trait analysis of genome-wide association summary statistics using MTAG. Nat. Genet. 50, 229–237 (2018).
Ilad, R. S., Fleming, S. D., Bebington, C. R. & Murphy, C. R. Ubiquitin is associated with the survival of ectopic stromal cells in endometriosis. Reprod. Biol. Endocrinol. 2, 69 (2004).
Cheng, J., Deng, Y. & Zhou, J. Role of the ubiquitin system in chronic pain. Front. Mol. Neurosci. 14, 674914 (2021).
Garcia-Gomez, E. et al. Regulation of inflammation pathways and inflammasome by sex steroid hormones in endometriosis. Front. Endocrinol. 10, 935 (2019).
Ding, Y. Q., Luo, H. & Qi, J. G. MHCII-restricted T helper cells: an emerging trigger for chronic tactile allodynia after nerve injuries. J. Neuroinflammation 17, 3 (2020).
Gougeon, A. Human ovarian follicular development: from activation of resting follicles to preovulatory maturation. Ann. Endocrinol. 71, 132–143 (2010).
Jones, M. R. & Goodarzi, M. O. Genetic determinants of polycystic ovary syndrome: progress and future directions. Fertil. Steril. 106, 25–32 (2016).
Ruth, K. S. et al. Genetic evidence that lower circulating FSH levels lengthen menstrual cycle, increase age at menopause and impact female reproductive health. Hum. Reprod. 31, 473–481 (2016).
Ruth, K. S. et al. Genome-wide association study with 1000 genomes imputation identifies signals for nine sex hormone-related phenotypes. Eur. J. Hum. Genet. 24, 284–290 (2016).
Gregus, A. M., Levine, I. S., Eddinger, K. A., Yaksh, T. L. & Buczynski, M. W. Sex differences in neuroimmune and glial mechanisms of pain. Pain 162, 2186–2200 (2021).
Lenert, M. E., Avona, A., Garner, K. M., Barron, L. R. & Burton, M. D. Sensory neurons, neuroimmunity, and pain modulation by sex hormones. Endocrinology 162, bqab109 (2021).
Choi, E. J. et al. Comorbidity of gynecological and non-gynecological diseases with adenomyosis and endometriosis. Obstet. Gynecol. Sci. 60, 579–586 (2017).
Loughlin, A. M. et al. Method used to identify adenomyosis and potentially undiagnosed adenomyosis in a large, U.S. electronic health record database. Pharmacoepidemiol. Drug Saf. 30, 1675–1686 (2021).
Powell, J. E. et al. Endometriosis risk alleles at 1p36.12 act through inverse regulation of CDC42 and LINC00339. Hum. Mol. Genet. 25, 5046–5058 (2016).
Cohen, S. P., Vase, L. & Hooten, W. M. Chronic pain: an update on burden, best practices, and new advances. Lancet 397, 2082–2097 (2021).
Kvaskoff, M. et al. Endometriosis: a high-risk population for major chronic diseases? Hum. Reprod. Update 21, 500–516 (2015).
Shafrir, A. L. et al. Co-occurrence of immune-mediated conditions and endometriosis among adolescents and adult women. Am. J. Reprod. Immunol. 86, e13404 (2021).
Tapmeier, T. T. et al. Neuropeptide S receptor 1 is a nonhormonal treatment target in endometriosis. Sci. Transl. Med. 13, eabd6469 (2021).
Shigesi, N. et al. The association between endometriosis and autoimmune diseases: a systematic review and meta-analysis. Hum. Reprod. Update 25, 486–503 (2019).
McGonagle, D. & McDermott, M. F. A proposed classification of the immunological diseases. PLoS Med. 3, e297 (2006).
Parazzini, F., Progretto Menopausa Italia Study Group Menopausal status, hormone replacement therapy use and risk of self-reported physician-diagnosed osteoarthritis in women attending menopause clinics in Italy. Maturitas 46, 207–212 (2003).
O’Mara, T. A., Spurdle, A. B. & Glubb, D. M., Endometrial Cancer Association Consortium Analysis of promoter-associated chromatin interactions reveals biologically relevant candidate target genes at endometrial cancer risk Loci. Cancers 11, 1440 (2019).
Genomes Project, C. et al. An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65 (2012).
Genomes Project, C. et al. A global reference for human genetic variation. Nature 526, 68–74 (2015).
McCarthy, S. et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet. 48, 1279–1283 (2016).
Jonsson, H. et al. Whole genome characterization of sequence diversity of 15,220 Icelanders. Sci. Data 4, 170115 (2017).
Mitt, M. et al. Improved imputation accuracy of rare and low-frequency variants using population-specific high-coverage WGS-based imputation reference panel. Eur. J. Hum. Genet. 25, 869–876 (2017).
Cook, J. P., Mahajan, A. & Morris, A. P. Guidance for the utility of linear models in meta-analysis of genetic association studies of binary phenotypes. Eur. J. Hum. Genet. 25, 240–245 (2017).
Devlin, B. & Roeder, K. Genomic control for association studies. Biometrics 55, 997–1004 (1999).
Willer, C. J., Li, Y. & Abecasis, G. R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191 (2010).
Magi, R., Lindgren, C. M. & Morris, A. P. Meta-analysis of sex-specific genome-wide association studies. Genet. Epidemiol. 34, 846–853 (2010).
Yang, J., Lee, S. H., Goddard, M. E. & Visscher, P. M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88, 76–82 (2011).
Wang, H. et al. SKAP-55 regulates integrin adhesion and formation of T cell-APC conjugates. Nat. Immunol. 4, 366–374 (2003).
Morris, A. P. et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat. Genet. 44, 981–990 (2012).
Janssen, E. B., Rijkers, A. C., Hoppenbrouwers, K., Meuleman, C. & D’Hooghe, T. M. Prevalence of endometriosis diagnosed by laparoscopy in adolescents with dysmenorrhea or chronic pelvic pain: a systematic review. Hum. Reprod. Update 19, 570–582 (2013).
Zondervan, K. T., Cardon, L. R. & Kennedy, S. H. The genetic basis of endometriosis. Curr. Opin. Obstet. Gynecol. 13, 309–314 (2001).
Wakefield, J. A Bayesian measure of the probability of false discovery in genetic epidemiology studies. Am. J. Hum. Genet. 81, 208–227 (2007).
Wellcome Trust Case Control, C. et al. Bayesian refinement of association signals for 14 loci in 3 common diseases. Nat. Genet. 44, 1294–1301 (2012).
Watanabe, K., Taskesen, E., van Bochoven, A. & Posthuma, D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 8, 1826 (2017).
O’Mara, T. A., Spurdle, A. B. & Glubb, D. M. Analysis of promoter-associated chromatin interactions reveals biologically relevant candidate target genes at endometrial cancer risk loci. Cancers 11, 1440 (2019).
Jain, A. & Tuteja, G. TissueEnrich: tissue-specific gene enrichment analysis. Bioinformatics 35, 1966–1967 (2019).
Uhlen, M. et al. Proteomics. Tissue-based map of the human proteome. Science 347, 1260419 (2015).
Consortium, G. T. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348, 648–660 (2015).
Fung, J. N. et al. The genetic regulation of transcription in human endometrial tissue. Hum. Reprod. 32, 893–904 (2017).
Gamazon, E. R. et al. Using an atlas of gene regulation across 44 human tissues to inform complex disease- and trait-associated variation. Nat. Genet. 50, 956–967 (2018).
Consortium, G. T. et al. Genetic effects on gene expression across human tissues. Nature 550, 204–213 (2017).
Qi, T. et al. Identifying gene targets for brain-related traits using transcriptomic and methylomic data from blood. Nat. Commun. 9, 2282 (2018).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Delaneau, O. et al. A complete tool set for molecular QTL discovery and analysis. Nat. Commun. 8, 15452 (2017).
Zhang, F. et al. OSCA: a tool for omic-data-based complex trait analysis. Genome Biol. 20, 107 (2019).
Eraslan, G. et al. Single-nucleus cross-tissue molecular reference maps toward understanding disease gene function. Science 376, eabl4290 (2022).
Gordon, M. L.T. forestplot: Advanced forest plot using ‘grid’ graphics. https://CRAN.R-project.org/package=forestplot (2017).
Hautakangas, H. et al. Genome-wide analysis of 102,084 migraine cases identifies 123 risk loci and subtype-specific risk alleles. Nat. Genet. 54, 152–160 (2022).
Johnston, K. J. A. et al. Genome-wide association study of multisite chronic pain in UK Biobank. PLoS Genet. 15, e1008164 (2019).
Acknowledgements
We thank all the study participants in the individual studies. We also thank many hospital directors and staff, gynecologists, general practitioners and pathology services who provided assistance with confirmation of diagnoses. We would like to thank the research participants and employees of 23andMe for making this work possible. We thank the women of the Icelandic deCODE study for their participation. We would like to express our gratitude to the staff and members of the Biobank Japan and Laboratory for Statistical Analysis, RIKEN Center for Integrative Medical Sciences for their outstanding assistance. We thank all the UK Biobank participants. Part of this research has been conducted using the UK Biobank Resource under application 9637. Generation Scotland received core support from the Chief Scientist Office of the Scottish Government Health Directorates (CZD/16/6) and the Scottish Funding Council (HR03006) and is currently supported by the Wellcome Trust (216767/Z/19/Z). Genotyping of the GS:SFH samples was carried out by the Genetics Core Laboratory at the Edinburgh Clinical Research Facility, University of Edinburgh, Scotland and was funded by the Medical Research Council UK and the Wellcome Trust (Wellcome Trust Strategic Award ‘STratifying Resilience and Depression Longitudinally’ (STRADL) reference 104036/Z/14/Z). The QIMR study was supported by grants from the National Health and Medical Research Council (NHMRC) of Australia (GNT241944, GNT339462, GNT389927, GNT389875, GNT389891, GNT389892, GNT389938, GNT443036, GNT442915, GNT442981, GNT496610, GNT496739, GNT552485, GNT552498, GNT1026033, GNT1050208 and GNT1147846), the Cooperative Research Centre for Discovery of Genes for Common Human Diseases (CRC), Cerylid Biosciences (Melbourne) and donations from N. Hawkins and S. Hawkins. Analyses of the QIMRHCS and OX GWAS were supported by the Wellcome Trust (WT084766/Z/08/Z) and made use of WTCCC2 control data generated by the Wellcome Trust Case-Control Consortium (awards 076113 and 085475). The ENDOX gene expression analyses were funded by the Medical Research Council UK (MR/K011480/1). We thank the participants of the Women’s Health Study: From Adolescence to Adulthood (A2A) for their valuable contributions and all staff of the Boston Center for Endometriosis. Financial support for the establishment of and data collection within the A2A cohort was provided by the J. Willard and Alice S. Marriott Foundation. A.L.S., K.L.T. and S.A.M. were supported by NICHD R01 HD094842 and NICHD R21 HD096358 from the Eunice Kennedy Shriver National Institute of Child Health and Human Development. We would like to thank the participants and staff of the Nurses’ Health Study (NHS) and Nurses’ Health Study 2 (NHS2) cohorts for their contributions. NHS/NHS2 are supported by grants UM1 CA186107 and UM1 CA176726 from the National Institutes of Health. C.T., P.K. and S.A.M. were supported by NICHD R01 HD096033. S.A.M. and K.T.Z. gratefully acknowledge funding provided by the Nezhat Family Foundation on behalf of Worldwide EndoMarch to their research programs. N.R. was supported by a grant from Wellbeing of Women UK (RG2031) and the EU Horizon 2020-funded project FEMaLe (ID 101017562). G.W.M. was supported by NHMRC Fellowships (GNT0613667, GNT1078399 and GNT1177194). D.R.N. was supported by NHMRC Fellowship (GNT0613674) and ARC Future Fellowship (FT0991022). S. Mortlock was supported by the Australian Government Medical Research Future Fund Research Grant (MRF1199785) and S. MacGregor by Australian National Health and Medical Research Council Fellowship. A.P.M. was supported in part by Versus Arthritis (grant 21754). The Melbourne study was supported by grants from the National Health and Medical Research Council (NHMRC) of Australia (GNT1026033, GNT1046880, GNT1049472, GNT1105321 and GNT1147846). R.M. and M.L. were supported by Estonian Research Council grant PRG1911, M.S. and M.P. by Estonian Research Council grant PRG1076 and A.S. by Estonian Research Council grant PRG1076, Horizon 2020 innovation (ERIN; EU952516) of the European Commission and Enter-prise Estonia (EU48695), MSCA-RISE-2020 project TRENDO (101008193). B.M., M.S.K., E.K., M.S., A.S.G. and D.S. were supported by Polish POIG grant 01.01.02-10-005/08 TESTOPLEK from the European Regional Development Fund. B.N. and D.W. were supported by NNF14CC0001, NNF17OC0027594 and NNF18SA0034956. J.F.D., J.E.G., M.H., S.H.C. and P.A.W.R. were supported by NHMRC GNT1105321. S.H., J.C.I., S.S., M.P., K.C.V., M.S. and L.C.G. were supported by NIH NICHD R01HD089511. J.K. was supported by The Sigrid Juselius Foundation, the Academy of Finland (297338 and 307247) and Novo Nordisk Foundation (NNF17OC0026062). C.M.L. was supported by the Li Ka Shing Foundation, NIHR Oxford Biomedical Research Centre, Oxford, NIH (1P50HD104224-01), Gates Foundation (INV-024200) and Wellcome Trust Investigator Award (221782/Z/20/Z). A.S. was supported by J. Willard and Alice S. Marriott Foundation, NIH NICHD R21 HD096358, NIH NICHD R01 HD094842. D.K.H. was supported by Wellbeing of Women (RG2137 and RG1073). C.H. was supported by MRC University Unit Programme Grant (MC_UU_00007/10—QTL in Health and Disease). D.C.W. was supported by NHMRC APP1155413, APP1185416, APP1073898 and APP1063061. M.N. acknowledge the Novo Nordisk Foundation (NNF21OC0071050). K.B. acknowledge the Novo Nordisk Foundation (NNF17OC0027594 and NNF14CC0001). TwinsUK receives funding from the Wellcome Trust (212904/Z/18/Z), Chronic Disease Research Foundation (CDRF) and European Union (H2020 contract 733100). TwinsUK and M.M. are supported by the National Institute for Health Research (NIHR)-funded BioResource, Clinical Research Facility and Biomedical Research Centre based at Guy’s and St Thomas’ NHS Foundation Trust in partnership with King’s College London.
Author information
Authors and Affiliations
Consortia
Contributions
N.R., A.P.M., S. Mortlock, M.G., P.L.M, L.S., M. Krassowski, M.L., M.N., D.P., E.S., N.F.T.S. and A.V. carried out data analysis. N.R., K.T.Z., A.P.M, G.W.M., S.A.M., M.N. and V.S. coordinated data analysis. N.R., S. Motlock, L.S., C.T., D.R.N., R.D., M.L., Y.S., P.C., S.S., A.G., K.B., D.W., G.T., B.M. and M.S. carried out cohort-level analyses. N.R., K.T.Z. and A.P.M. coordinated cohort-level analyses. N.R., K.T.Z. and A.P.M. drafted the manuscript. N.R., S. Mortlock, M.G., P.L.M., L.S., G.G., C.T., R.D., M.H.L., Y.S., P.C., S.S., A.G., K.B., M. Krassowski, M.L., B.M., M.N., D.P., E.S., M.S.K., G.T., A.V., D.W., R.A., K.S.B., A.C., C.S.K.C., C.C., J.C., I.D., A.D., D.O., J.F.D., T.E., P.F., J.N.F, R.T.G., J.G., P.H., H.R.H., M.H., O.H., S.H.C., I.C.H., H.H., S.H., J.C.I., M.R.J., Y.K., S.H.K., E.K., J.K., M. Kubo, B.K., V.K., H.L., M.R.L, C.L.M., S. MacGregor, M.M., N.G.M., C.M., M.M., A.D.M, A.N., C.N., C.M.O., J.O.A., S.P., M. Paranjpe, M. Peters, G.P., D.J.P., J.R., K.M.R., H.R., M.S., L.S., A.J.S., S.S., A.S., A.S.G., M.S., B.H.S, B.S., T.S., K.S., A.T., K.L.T., C.T., S.A.T., A.V., K.V., K.C.V., D.J.W., E.Z., M.I.Z, S.A., J.E.B, P.M.R., T.D., G.N.G., D.K.H., C.H., A.W.H., S.K.L., H.M., D.I.C., P.A.W.R., P.T.S., M.S., T.S., D.S., J.Y.T., D.C.W., L.C.G., D.R.V.E., O.U., P.K., A.S., D.R.N., R.M., K.S., C.M.B., P.Y.B., V.S., M.N., S.A.M., G.W.M., A.P.M. and K.T.Z. provided and/or processed phenotype and/or genotype data. All authors contributed and discussed the results and commented on the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
L.S., G.T., K.S. and V.S. are employees of deCODE Genetics/Amgen. G.G., C.C. and P.Y.B. are employees of Celmatix Inc. and hold stock or stock options in Celmatix Inc. P.F. and J.Y.T. are employees of 23andMe Inc. O.H. serves occasionally on advisory boards for Bayer AG, Gedeon Richter, HRA Pharma, and Vifor Pharma and has lectured at educational events for Bayer AG, Gedeon Richter, and Sandoz AG. C.M.L. receives grant support from Bayer AG, Novo Nordisk and her husband works for Vertex. J.R.’s spouse receives salary and stock options from Merck & Co. K.V. receives research funding from Bayer Healthcare and Honorarium for Consultancy and Lectures from Bayer Healthcare, Grunenthal GmBH, AbbVie and Eli Lilly. T.D. is an employee of Merck Healthcare KGaA, Darmstadt, Germany. T.S. is the co-founder and shareholder of Zoe Global Ltd. P.A.W.R. is an advisory board member for Bayer AG. O.U. lectured at educational events for Exeltis and Merck. K.L.T. report an endometriosis grant from Aspira, which is not relevant to the submitted work. K.T.Z. and C.M.B. reported grants from Bayer A.G., AbbVie Inc., Volition Rx, MDNA Life Sciences, Roche Diagnostics Inc. outside the submitted work. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Genetics thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Q-Q plots for genome-wide association results.
Q-Q plot for genome-wide association results for a. overall endometriosis, b. rASRM stage I/II endometriosis, c. rASRM stage III/IV endometriosis, d. endometriosis-associated infertility, e. only studies with >300 cases.
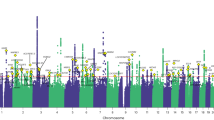
Extended Data Fig. 2 Manhattan plots for genome-wide association results.
Manhattan plot for genome-wide association results for a. overall endometriosis, b. rASRM stage I/II endometriosis, c. rASRM stage III/IV endometriosis, d. endometriosis associated infertility. The GWAS meta-analysis results are shown on the y-axis as –log10(P-value) and on the x-axis is the chromosomal location. The red vertical line illustrates the genome-wide significance (p < 5 × 10−8) and the blue vertical line shows the nominal genome-wide results (p < 1 × 10−5).
Extended Data Fig. 3 Comparison of mixed female and male controls GWAS meta-analysis vs. female only controls GWAS meta-analysis results.
Results from meta-analysis contrasting GWAS studies with mixed female and male controls (N cases=5,222, N controls=44,176) vs. GWAS studies with only female controls (N cases=44,176, N controls=657,747). Significant heterogeneity: p-value<1.19x10−3 (0.05/42). The error bars represent standard error estimates for the beta co-efficient estimates. * SNPs with nominal heterogeneity p-values: rs1430787, p = 0.03, rs1451383, p = 0.01.
Extended Data Fig. 4 Tissue-specific gene enrichment results.
Tissue-specific gene enrichment using RNA-Seq data across 35 human tissues from the Human Protein Atlas (a and b) and 29 human tissues from GTEx (c and d). The x-axis shows each of the tissues, and the y-axis represents the tissue-specific gene enrichment −Log10(P−Value) (left) and the fold-change values of the tissue-specific gene enrichment (right).
Extended Data Fig. 5 SKAP1/17q21.32 locus.
a. Illustration of the regional association plot for the SKAP1/17q21.32 locus including the 99% credible sets. eSMR endometrium SNP: SMR SNP identified as causal for endometriosis utilizing the eQTL data from endometrium; The shaded region in the credible sets panel is further annotated in panel c. b. SMR significant endometrial eQTL for HOXB9 (SMR p-value=2x10−6). The lower and upper bounds of the boxes represent the first and third quantiles, the whiskers extend 1.5 times the interquartile range from the bounds of the box and the line represents the median. c. position of the SMR-significant eQTL in the SKAP1/17q21.32 locus along with HOXB9 promoter-associated chromatin loops with anchor points containing the lead SNP and the SMR SNP. The SMR-significant SNP associated with expression of HOXB9 in endometrium (eQTL) and endometriosis is shown in black and the associated HOXB9 target is shown in red. Valid promoter-associated chromatin loops were generated from H3K27Ac HiChIP libraries from a normal immortalized endometrial cell line (E6E7hTERT) and three endometrial cancer cell lines (ARK1, Ishikawa and JHUEM-14)68.
Extended Data Fig. 6 sICAM-1 level results.
Box plots of log scale sICAM-1 levels in a. overall endometriosis cases (N=136) vs. controls (N=54), b. rASRM stage I/II cases (N=85) vs. controls and rASRM stage III/IV (N=51) vs. controls. Reported p-values are from the adjusted logistic regression model (see Supplementary Information: Methods) (Supplementary Table 19). The lower and upper bounds of the boxes represent the first and third quantiles, the whiskers extend 1.5 times the interquartile range from the bounds of the box and the line represents the median.
Extended Data Fig. 7 Single cell expression profiles.
Single cell expression profiles for 18/23 genes regulated by endometriosis risk variants in GTEx Multi-Gene Single Cell Viewer (5 were not included in the database). This is an aster plot where the fraction of cells in which a gene is detected is shown. The cells are categorized by cell-type.
Extended Data Fig. 8 Correlation of 42 GWAS loci between endometriosis surgical sub-types and adenomyosis.
Correlation between the effect sizes of 42 endometriosis-associated GWAS loci contrasting endometriosis surgical sub-types and adenomyosis: a. Adenomyosis vs. rASRM stage I/II, b. Adenomyosis vs. rASRM stage III/IV, c. Adenomyosis vs. endometrioma, d. Adenomyosis vs. deep lesions, e. Adenomyosis vs. superficial lesions. Minor allele frequency for each of the 42 variants is given by shade of gray: Lighter shade of gray designates a smaller MAF, darker shade of gray a larger MAF. Nominal associations (p < 0.05) are annotated with locus name and larger circles. The solid black line represents the linear regression line and the dotted black line is the x=y with a slope of 1 for reference of change in ORs. Test statistics including p-values for all the associations are provided in Supplementary Table 21.
Extended Data Fig. 9 Genetic correlation results between only surgically or medically confirmed endometriosis and 32 traits/conditions.
Genetic correlation between only surgically or medically confirmed endometriosis (N=8,390 cases) and 32 immune/inflammatory, pain, reproductive, and metabolic traits/conditions using LD score regression analysis (LDSC). Heritability of each trait is noted in parenthesis on the x-axis. The significance threshold is adjusted for multiple testing using Bonferroni correction. Significant (p < 1.56 × 10−3) correlations are denoted with a red star (*), nominal correlation (p < 0.05) with a green star (*). Bars present the genetic correlation (rg) for each trait in relation to endometriosis and the error bars are standard errors. The exact p-values are provided in Supplementary Table 25.
Extended Data Fig. 10 Genetic correlation results between endometriosis and 19 brain imaging traits in UKBB.
Genetic correlation between 19 brain imaging traits in UKBB and a. endometriosis (N=58,961) and b. surgically or medically confirmed endometriosis (N=8,390 cases) using LD score regression analysis (LDSC). A total of 6 functional MRI measures (netmat_ICA_09Aug2017_001-006), structural MRI measures including 11 freesurfer derived variables (aseg_lh_volume_Left-Thalamus-Proper, aseg_lh_volume_Left-Hippocampus, aseg_lh_ volume_Left-Amygdala, aseg_lh_volume_Right-Thalamus-Proper, aseg_lh_volume_Right-Hippocampus, aseg_lh_volume_Right-Amygdala, aseg_lh_volume_CC_Posterior, aseg_lh_volume_CC_Mid_Posterior, aseg_lh_volume_CC_Central, aseg_lh_volume_CC_Mid_Anterior, aseg_lh_volume_CC_Anterior) and 2 FAST calculations for insula region (IDP_T1_FAST_ROIs_L_insular_cortex, IDP_T1_FAST_ROIs_R_insular_cortex) were analyzed. Heritability of each trait is noted in parenthesis on the x-axis. Nominal correlations are denoted with a green star (*). Bars present the genetic correlation (rg) for each trait in relation to endometriosis and the error bars are standard errors. The exact p-values are provided in Supplementary Tables 27 and 28.
Supplementary information
Supplementary Information
Supplementary Text, Supplementary Methods, Contributors to FinnGen, Contributors to DBDS Genomic Consortium, Ethics Statements and Supplementary Figs. 1–8.
Supplementary Tables
Supplementary Tables 1–35.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Rahmioglu, N., Mortlock, S., Ghiasi, M. et al. The genetic basis of endometriosis and comorbidity with other pain and inflammatory conditions. Nat Genet 55, 423–436 (2023). https://doi.org/10.1038/s41588-023-01323-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-023-01323-z
This article is cited by
-
Quality of life and symptoms of pain in patients with endometriomas compared to those with other endometriosis lesions: a cross-sectional study
BMC Women's Health (2024)
-
Causal effects of endometriosis on SLE, RA and SS risk: evidence from meta-analysis and Mendelian randomization
BMC Pregnancy and Childbirth (2024)
-
Surge in endometriosis research after decades of underfunding could herald new era for women’s health
Nature Medicine (2024)
-
Impaired bone morphogenetic protein (BMP) signaling pathways disrupt decidualization in endometriosis
Communications Biology (2024)
-
A life-course approach to women’s health
Nature Medicine (2024)