Abstract
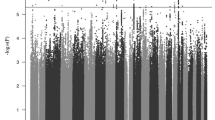
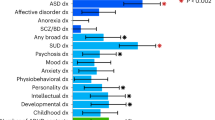
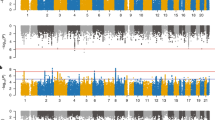
Attention-deficit hyperactivity disorder (ADHD) is a neurodevelopmental disorder with onset in childhood (childhood ADHD); two-thirds of affected individuals continue to have ADHD in adulthood (persistent ADHD), and sometimes ADHD is diagnosed in adulthood (late-diagnosed ADHD). We evaluated genetic differences among childhood (n = 14,878), persistent (n = 1,473) and late-diagnosed (n = 6,961) ADHD cases alongside 38,303 controls, and rare variant differences in 7,650 ADHD cases and 8,649 controls. We identified four genome-wide significant loci for childhood ADHD and one for late-diagnosed ADHD. We found increased polygenic scores for ADHD in persistent ADHD compared with the other two groups. Childhood ADHD had higher genetic overlap with hyperactivity and autism compared with late-diagnosed ADHD and the highest burden of rare protein-truncating variants in evolutionarily constrained genes. Late-diagnosed ADHD had a larger genetic overlap with depression than childhood ADHD and no increased burden in rare protein-truncating variants. Overall, these results suggest a genetic influence on age at first ADHD diagnosis, persistence of ADHD and the different comorbidity patterns among the groups.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
Summary statistics from GWAS of childhood, persistent and late-diagnosed ADHD are available at the iPSYCH website (https://ipsych.dk/en/research/downloads/). All relevant iPSYCH data are available from the authors after approval by the iPSYCH Data Access Committee and can only be accessed on the secure Danish server (GenomeDK; https://genome.au.dk) as the data are protected by Danish legislation. For data access, please contact: D.D. or A.D.B. (anders@biomed.au.dk). Correspondence and requests for materials should be addressed to D.D. (ditte@biomed.au.dk).
Code availability
No previously unreported custom computer code or algorithm were used to generate results, all software used in the study are publicly available from the Internet as described in Methods and Reporting Summary.
References
Faraone, S. et al. Attention-deficit/hyperactivity disorder. Nat. Rev. Dis. Primers 1, 15020 (2015).
Franke, B. et al. The genetics of attention deficit/hyperactivity disorder in adults, a review. Mol. Psychiatry 17, 960–987 (2012).
Faraone, S. V. & Larsson, H. Genetics of attention deficit hyperactivity disorder. Mol. Psychiatry 24, 562–575 (2019).
Demontis, D. et al. Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat. Genet. 51, 63–75 (2019).
Faraone, S. V., Biederman, J. & Mick, E. The age-dependent decline of attention deficit hyperactivity disorder: a meta-analysis of follow-up studies. Psychol. Med. 36, 159–165 (2006).
Fatseas, M. et al. Addiction severity pattern associated with adult and childhood attention deficit hyperactivity disorder (ADHD) in patients with addictions. Psychiatry Res 246, 656–662 (2016).
Agnew-Blais, J. C. et al. Young adult mental health and functional outcomes among individuals with remitted, persistent and late-onset ADHD. Br. J. Psychiatry 213, 526–534 (2018).
Ilbegi, S. et al. Substance use and nicotine dependence in persistent, remittent, and late-onset ADHD: a 10-year longitudinal study from childhood to young adulthood. J. Neurodev. Disord. 10, 42 (2018).
Yoshimasu, K. et al. Adults with persistent ADHD: gender and psychiatric comorbidities - a population-based longitudinal study. J. Atten. Disord. 22, 535–546 (2018).
Kim, K. M. et al. Psychopathological, temperamental, and characteristic factors in adults with remaining childhood attention-deficit hyperactivity symptoms. Int J. Psychiatry Clin. Pract. 21, 236–241 (2017).
Tandon, M., Tillman, R., Agrawal, A. & Luby, J. Trajectories of ADHD severity over 10 years from childhood into adulthood. Atten. Defic. Hyperact Disord. 8, 121–130 (2016).
Kan, K. J. et al. Genetic and environmental stability in attention problems across the lifespan: evidence from the Netherlands twin register. J. Am. Acad. Child Adolesc. Psychiatry 52, 12–25 (2013).
Larsson, H. et al. Genetic and environmental influences on adult attention deficit hyperactivity disorder symptoms: a large Swedish population-based study of twins. Psychol. Med. 43, 197–207 (2013).
Brikell, I., Kuja-Halkola, R. & Larsson, H. Heritability of attention-deficit hyperactivity disorder in adults. Am. J. Med. Genet. B Neuropsychiatr. Genet.168, 406–413 (2015).
Pingault, J. B. et al. Genetic and environmental influences on the developmental course of attention-deficit/hyperactivity disorder symptoms from childhood to adolescence. JAMA Psychiatry 72, 651–658 (2015).
Riglin, L. et al. Association of genetic risk variants with attention-deficit/hyperactivity disorder trajectories in the general population. JAMA Psychiatry 73, 1285–1292 (2016).
Asherson, P. & Agnew-Blais, J. Annual research review: does late-onset attention-deficit/hyperactivity disorder exist? J. Child Psychol. Psychiatry 60, 333–352 (2019).
Agnew-Blais, J. C. et al. Polygenic risk and the course of attention-deficit/hyperactivity disorder from childhood to young adulthood: findings from a nationally-representative cohort. J. Am. Acad. Child Adolesc. Psychiatry 60, 1147–1156 (2021).
Rovira, P. et al. Shared genetic background between children and adults with attention deficit/hyperactivity disorder. Neuropsychopharmacology 45, 1617–1626 (2020).
Pedersen, C. B. et al. The iPSYCH2012 case-cohort sample: new directions for unravelling genetic and environmental architectures of severe mental disorders. Mol. Psychiatry 23, 6–14 (2018).
Yang, J., Lee, S. H., Goddard, M. E. & Visscher, P. M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88, 76–82 (2011).
Zayats, T. QIMR Group, SickKids Group, TEDS Group, CATSS Group, MoBa Group, NTR Group, ALSPAC Group, ATGU Group. Genome-wide association meta-analysis of ADHD symptomatology: inattention and hyperactivity/impulsivity. Eur. Neuropsychopharmacol. 29, S1190 (2019).
Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427 (2014).
Stahl, E. A. et al. Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat. Genet. 51, 793–803 (2019).
Howard, D. M. et al. Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat. Neurosci. 22, 343–352 (2019).
Grove, J. et al. Identification of common genetic risk variants for autism spectrum disorder. Nat. Genet. 51, 431–444 (2019).
Watson, H. J. et al. Genome-wide association study identifies eight risk loci and implicates metabo-psychiatric origins for anorexia nervosa. Nat. Genet. 51, 1207–1214 (2019).
International Obsessive Compulsive Disorder Foundation Genetics Collaborative (IOCDF-GC) & OCD Collaborative Genetics Association Studies (OCGAS) Revealing the complex genetic architecture of obsessive-compulsive disorder using meta-analysis. Mol. Psychiatry 23, 1181–1188 (2018).
Johnson, E. C. et al. A large-scale genome-wide association study meta-analysis of cannabis use disorder. Lancet Psychiatry 7, 1032–1045 (2020).
Walters, R. K. et al. Transancestral GWAS of alcohol dependence reveals common genetic underpinnings with psychiatric disorders. Nat. Neurosci. 21, 1656–1669 (2018).
Lee, J. J. et al. Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat. Genet. 50, 1112–1121 (2018).
Yengo, L. et al. Meta-analysis of genome-wide association studies for height and body mass index in approximately 700000 individuals of European ancestry. Hum. Mol. Genet. 27, 3641–3649 (2018).
Barban, N. et al. Genome-wide analysis identifies 12 loci influencing human reproductive behavior. Nat. Genet. 48, 1462–1472 (2016).
Watanabe, K. et al. A global overview of pleiotropy and genetic architecture in complex traits. Nat. Genet. 51, 1339–1348 (2019).
Jansen, P. R. et al. Genome-wide analysis of insomnia in 1,331,010 individuals identifies new risk loci and functional pathways. Nat. Genet. 51, 394–403 (2019).
Satterstrom, F. K. et al. Autism spectrum disorder and attention deficit hyperactivity disorder have a similar burden of rare protein-truncating variants. Nat. Neurosci. 22, 1961–1965 (2019).
Lek, M. et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 (2016).
Kaplanis, J. et al. Evidence for 28 genetic disorders discovered by combining healthcare and research data. Nature 586, 757–762 (2020).
Cross-Disorder Group of the Psychiatric Genomics Consortium. Genomic relationships, novel loci, and pleiotropic mechanisms across eight psychiatric disorders. Cell 179, 1469–1482.e11 (2019).
Enard, W. et al. Molecular evolution of FOXP2, a gene involved in speech and language. Nature 418, 869–872 (2002).
Schreiweis, C. et al. Humanized Foxp2 accelerates learning by enhancing transitions from declarative to procedural performance. Proc. Natl Acad. Sci. USA 111, 14253–14258 (2014).
Vernes, S. C. et al. Foxp2 regulates gene networks implicated in neurite outgrowth in the developing brain. PLoS Genet. 7, e1002145 (2011).
Rommelse, N. N., Franke, B., Geurts, H. M., Hartman, C. A. & Buitelaar, J. K. Shared heritability of attention-deficit/hyperactivity disorder and autism spectrum disorder. Eur. Child Adolesc. Psychiatry 19, 281–295 (2010).
Skirrow, C. & Asherson, P. Emotional lability, comorbidity and impairment in adults with attention-deficit hyperactivity disorder. J. Affect. Disord. 147, 80–86 (2013).
Chang, Z., D’Onofrio, B. M., Quinn, P. D., Lichtenstein, P. & Larsson, H. Medication for attention-deficit/hyperactivity disorder and risk for depression: a nationwide longitudinal cohort study. Biol. Psychiatry 80, 916–922 (2016).
Biederman, J., Petty, C. R., Evans, M., Small, J. & Faraone, S. V. How persistent is ADHD? A controlled 10-year follow-up study of boys with ADHD. Psychiatry Res. 177, 299–304 (2010).
Van Veen, M. M., Kooij, J. J., Boonstra, A. M., Gordijn, M. C. & Van Someren, E. J. Delayed circadian rhythm in adults with attention-deficit/hyperactivity disorder and chronic sleep-onset insomnia. Biol. Psychiatry 67, 1091–1096 (2010).
Pedersen, C. B., Gotzsche, H., Moller, J. O. & Mortensen, P. B. The Danish civil registration system. A cohort of eight million persons. Dan. Med. Bull. 53, 441–449 (2006).
Mors, O., Perto, G. P. & Mortensen, P. B. The Danish Psychiatric Central Research Register. Scand. J. Public Health 39, 54–57 (2011).
Nørgaard-Pedersen, B. & Hougaard, D. M. Storage policies and use of the Danish Newborn Screening Biobank. J. Inherit. Metab. Dis. 30, 530–536 (2007).
Pedersen, C. B. The Danish civil registration system. Scand. J. Public Health 39, 22–25 (2011).
Borglum, A. D. et al. Genome-wide study of association and interaction with maternal cytomegalovirus infection suggests new schizophrenia loci. Mol. Psychiatry 19, 325–333 (2014).
Hollegaard, M. V. et al. Robustness of genome-wide scanning using archived dried blood spot samples as a DNA source. BMC Genet. 12, 58 (2011).
Illumina GenCall Data Analysis Software Illumina Tech Note (Illumina, 2005).
Korn, J. M. et al. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat. Genet. 40, 1253–1260 (2008).
Iglesias, A. I. et al. Haplotype reference consortium panel: practical implications of imputations with large reference panels. Hum. Mutat. 38, 1025–1032 (2017).
Loh, P. R. et al. Reference-based phasing using the haplotype reference consortium panel. Nat. Genet. 48, 1443–1448 (2016).
Das, S. et al. Next-generation genotype imputation service and methods. Nat. Genet. 48, 1284–1287 (2016).
Price, A. L. et al. Long-range LD can confound genome scans in admixed populations. Am. J. Hum. Genet. 83, 132–135 (2008); author reply, 135–139.
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Chang, C. C. et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4, 7 (2015).
Price, A. L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006).
Galinsky, K. J. et al. Fast principal-component analysis reveals convergent evolution of ADH1B in Europe and East Asia. Am. J. Hum. Genet. 98, 456–472 (2016).
Dalsgaard, S. et al. Incidence rates and cumulative incidences of the full spectrum of diagnosed mental disorders in childhood and adolescence. JAMA Psychiatry 77, 155–164 (2020).
Bulik-Sullivan, B. K. et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47, 291–295 (2015).
Hubel, C. et al. Genomics of body fat percentage may contribute to sex bias in anorexia nervosa. Am. J. Med. Genet. B Neuropsychiatr. Genet.180, 428–438 (2019).
Poulsen, J. B. et al. High-quality exome sequencing of whole-genome amplified neonatal dried blood spot DNA. PLoS ONE 11, e0153253 (2016).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Van der Auwera, G. A. et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinformatics 43, 11.10.1–11.10.33 (2013).
Kalia, S. S. et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): a policy statement of the American College of Medical Genetics and Genomics. Genet. Med. 19, 249–255 (2017).
Cingolani, P. et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 6, 80–92 (2012).
Karczewski, K. J. et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443 (2020).
Acknowledgements
D.D. was supported by the Novo Nordisk Foundation (NNF20OC0065561) and the Lundbeck Foundation (R344-2020-1060). The iPSYCH team was supported by grants from the Lundbeck Foundation (R102-A9118, R155-2014-1724 and R248-2017-2003), the EU FP7 Program (grant number 602805, ‘Aggressotype’) and H2020 Program (grant number 667302, ‘CoCA’), National Institute of Mental Health (1U01MH109514-01 to A.D.B.) and the universities and university hospitals of Aarhus and Copenhagen. The Danish National Biobank resource was supported by the Novo Nordisk Foundation. High-performance computer capacity for handling and statistical analysis of iPSYCH data at the GenomeDK high-performance computer facility was provided by the Center for Genomics and Personalized Medicine and the Centre for Integrative Sequencing, iSEQ, Aarhus University, Denmark (grant to A.D.B.). M.S.A. is a recipient of a Juan de la Cierva Incorporación contract from the Ministry of Science, Innovation and Universities, Spain (IJC2018-035346-I). The research leading to these results has received funding from the Instituto de Salud Carlos III (PI17/00289, PI18/01788, P19/01224 and PI20/00041) and from the Agència de Gestió d’Ajuts Universitaris i de Recerca-AGAUR, Generalitat de Catalunya (2017SGR1461) and was cofinanced by the European Regional Development Fund. The Norwegian Mother, Father and Child Cohort Study is supported by the Norwegian Ministry of Health and Care Services and the Ministry of Education and Research. We are grateful to all the families in Norway who have taken part in this ongoing cohort study. L.V.R. is a recipient of a predoctoral fellowship from the Instituto de Salud Carlos III, Spain (FI18/00285). M.R. was a recipient of a Miguel de Servet contract from the Instituto de Salud Carlos III, Spain (CP09/00119 and CPII15/00023). T.Z. is funded by R37MH107649-07S1 and by the Research Council of Norway (grant number 288083).
Author information
Authors and Affiliations
Contributions
V.M.R, J.D. and L.V.-R. carried out the analysis. J.G., T. Z., J.A.R.-Q., F.K.S., M.S.A., J.B.-G., M.B.-H., T.D.A., A.R., M.J.D., B.M.N., M.N., T.W., O.M., D.M.H. and P.B.M. performed sample and/or data provision and processing. D.D. and V.M.R. wrote the manuscript. D.D., V.M.R., F.K.S., A.D.B. and M.R. revised the manuscript. D.D. and V.M.R. were responsible for the study design. All authors contributed with critical revisions of the manuscript.
Corresponding author
Ethics declarations
Competing interests
D.D. has received a speaker fee from Takeda. J.A.R.-Q. has been on the speaker’s bureau and/or has acted as a consultant for Janssen-Cilag, Novartis, Shire, Takeda, Bial, Shionogi, Sincrolab, Novartis, BMS, Medice and Rubiand Raffo in the past 3 years. He has also received travel awards (air tickets and hotel) for taking part in psychiatric meetings from Janssen-Cilag, RubiShire, Takeda, Shionogi, Bial and Medice. The Department of Psychiatry chaired by him has received unrestricted educational and research support from the following companies in the past 3 years: Janssen-Cilag, Shire, Oryzon, Roche, Psious and Rubió. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Genetics thanks the anonymous reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–7, Tables 1–12 and Notes 1–3.
Supplementary Table 1
Supplementary Tables 1–12
Rights and permissions
About this article
Cite this article
Rajagopal, V.M., Duan, J., Vilar-Ribó, L. et al. Differences in the genetic architecture of common and rare variants in childhood, persistent and late-diagnosed attention-deficit hyperactivity disorder. Nat Genet 54, 1117–1124 (2022). https://doi.org/10.1038/s41588-022-01143-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-022-01143-7
This article is cited by
-
Genetic architecture and biology of youth-onset type 2 diabetes
Nature Metabolism (2024)
-
Attention-deficit/hyperactivity disorder
Nature Reviews Disease Primers (2024)
-
Polygenic profiles define aspects of clinical heterogeneity in attention deficit hyperactivity disorder
Nature Genetics (2024)
-
Neurogenetic mechanisms of risk for ADHD: Examining associations of polygenic scores and brain volumes in a population cohort
Journal of Neurodevelopmental Disorders (2023)
-
Selecting cases of major psychiatric and substance use disorders in Swedish national registries on the basis of clinical features to maximize the strength or specificity of the genetic risk
Molecular Psychiatry (2023)