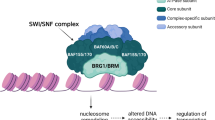
Abstract
Mammalian SWI/SNF (mSWI/SNF) ATP-dependent chromatin remodeling complexes establish and maintain chromatin accessibility and gene expression, and are frequently perturbed in cancer. Clear cell meningioma (CCM), an aggressive tumor of the central nervous system, is uniformly driven by loss of SMARCE1, an integral subunit of the mSWI/SNF core. Here, we identify a structural role for SMARCE1 in selectively stabilizing the canonical BAF (cBAF) complex core–ATPase module interaction. In CCM, cBAF complexes fail to stabilize on chromatin, reducing enhancer accessibility, and residual core module components increase the formation of BRD9-containing non-canonical BAF (ncBAF) complexes. Combined attenuation of cBAF function and increased ncBAF complex activity generates the CCM-specific gene expression signature, which is distinct from that of NF2-mutated meningiomas. Importantly, SMARCE1-deficient cells exhibit heightened sensitivity to small-molecule inhibition of ncBAF complexes. These data inform the function of a previously elusive SWI/SNF subunit and suggest potential therapeutic approaches for intractable SMARCE1-deficient CCM tumors.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
All genomic data contained in this manuscript have been deposited on the Gene Expression Omnibus (GEO) repository, https://www.ncbi.nlm.nih.gov/geo/, under accession code GSE174360. Source data are provided with this paper or are provided as Supplementary Information. Raw sequencing data from the fresh-frozen primary meningioma tumor samples (controlled access samples) can be made available upon request to C. Kadoch and D. Meredith, with a time frame for response of 1–2 days, and data availability within 2–3 weeks.
Code availability
No custom code was generated for this study.
References
Clapier, C. R., Iwasa, J., Cairns, B. R. & Peterson, C. L. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat. Rev. Mol. Cell Biol. 18, 407–422 (2017).
Mashtalir, N. et al. Modular organization and assembly of SWI/SNF family chromatin remodeling complexes. Cell 175, 1272–1288 e20 (2018).
Michel, B. C. et al. A non-canonical SWI/SNF complex is a synthetic lethal target in cancers driven by BAF complex perturbation. Nat. Cell Biol. 20, 1410–1420 (2018).
Kadoch, C. et al. Proteomic and bioinformatic analysis of mSWI/SNF (BAF) complexes reveals extensive roles in human malignancy. Nat. Genet. 45, 592–601 (2013).
Garraway, L. A. & Lander, E. S. Lessons from the cancer genome. Cell 153, 17–37 (2013).
Pulice, J. L. & Kadoch, C. Composition and function of mammalian SWI/SNF chromatin remodeling complexes in human disease. Cold Spring Harb. Symp. Quant. Biol. 81, 53–60 (2016).
Versteege, I. et al. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature 393, 203–206 (1998).
Biegel, J. A. et al. Germ-line and acquired mutations of INI1 in atypical teratoid and rhabdoid tumors. Cancer Res. 59, 74–79 (1999).
Storlazzi, C. T. et al. A novel fusion gene, SS18L1/SSX1, in synovial sarcoma. Genes Chromosomes Cancer 37, 195–200 (2003).
Ramos, P. et al. Small cell carcinoma of the ovary, hypercalcemic type, displays frequent inactivating germline and somatic mutations in SMARCA4. Nat. Genet. 46, 427–429 (2014).
Jones, S. et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science 330, 228–231 (2010).
Sauter, J. L. et al. SMARCA4-deficient thoracic sarcoma: a distinctive clinicopathological entity with undifferentiated rhabdoid morphology and aggressive behavior. Mod. Pathol. 30, 1422–1432 (2017).
Cao, S. et al. Comprehensive characterization of cancer driver genes and mutations. Cell 174, 1034–1035 (2018).
Pan, J. et al. The ATPase module of mammalian SWI/SNF family complexes mediates subcomplex identity and catalytic activity-independent genomic targeting. Nat. Genet. 51, 618–626 (2019).
Mashtalir, N. et al. A structural model of the endogenous human BAF complex informs disease mechanisms. Cell 183, 802–817.e24 (2020).
Nakayama, R. T. et al. SMARCB1 is required for widespread BAF complex-mediated activation of enhancers and bivalent promoters. Nat. Genet. 49, 1613–1623 (2017).
Wang, X. et al. SMARCB1-mediated SWI/SNF complex function is essential for enhancer regulation. Nat. Genet. 49, 289–295 (2017).
Mathur, R. et al. ARID1A loss impairs enhancer-mediated gene regulation and drives colon cancer in mice. Nat. Genet. 49, 296–302 (2017).
Kelso, T. W. R. et al. Chromatin accessibility underlies synthetic lethality of SWI/SNF subunits in ARID1A-mutant cancers. eLife 6, e30506 (2017).
Smith, M. J. et al. Loss-of-function mutations in SMARCE1 cause an inherited disorder of multiple spinal meningiomas. Nat. Genet. 45, 295–298 (2013).
Smith, M. J. et al. Germline SMARCE1 mutations predispose to both spinal and cranial clear cell meningiomas. J. Pathol. 234, 436–440 (2014).
Gerkes, E. H. et al. A heritable form of SMARCE1-related meningiomas with important implications for follow-up and family screening. Neurogenetics 17, 83–89 (2016).
Tauziede-Espariat, A. et al. Loss of SMARCE1 expression is a specific diagnostic marker of clear cell meningioma: a comprehensive immunophenotypical and molecular analysis. Brain Pathol. 28, 466–474 (2018).
Inoue, T. et al. Hereditary clear cell meningiomas in a single family: three-cases report. Acta Neurochir. (Wien.) 160, 2321–2325 (2018).
Libert, D. M. & Prayson, R. A. Pediatric clear cell meningioma involving the middle cranial fossa in the context of NF2 and SMARCE1 mutations. Ann. Diagn. Pathol. 46, 151516 (2020).
Shoakazemi, A. et al. The importance of genetic counseling and screening for people with pathogenic SMARCE1 variants: a family study. Am. J. Med. Genet. A 185, 561–565 (2021).
Navalkele, P., Guzman, M., Kirby, A., Pinz, H. & Kemp, J. Familial pediatric clear cell meningioma with germline SMARCE1 mutation in the United States. J. Neuropathol. Exp. Neurol. 79, 1250–1252 (2020).
Sievers, P. et al. Clear cell meningiomas are defined by a highly distinct DNA methylation profile and mutations in SMARCE1. Acta Neuropathol. 141, 281–290 (2021).
Smith, M. J. Germline and somatic mutations in meningiomas. Cancer Genet. 208, 107–114 (2015).
Clark, V. E. et al. Genomic analysis of non-NF2 meningiomas reveals mutations in TRAF7, KLF4, AKT1, and SMO. Science 339, 1077–1080 (2013).
Abedalthagafi, M. et al. Oncogenic PI3K mutations are as common as AKT1 and SMO mutations in meningioma. Neuro Oncol. 18, 649–655 (2016).
Bi, W. L., Prabhu, V. C. & Dunn, I. F. High-grade meningiomas: biology and implications. Neurosurg. Focus 44, E2 (2018).
Ye, Y. et al. Structure of the RSC complex bound to the nucleosome. Science 366, 838–843 (2019).
He, S. et al. Structure of nucleosome-bound human BAF complex. Science 367, 875–881 (2020).
Han, Y., Reyes, A. A., Malik, S. & He, Y. Cryo-EM structure of SWI/SNF complex bound to a nucleosome. Nature 579, 452–455 (2020).
Wagner, F. R. et al. Structure of SWI/SNF chromatin remodeller RSC bound to a nucleosome. Nature 579, 448–451 (2020).
Estruch, F. & Carlson, M. SNF6 encodes a nuclear protein that is required for expression of many genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 10, 2544–2553 (1990).
Cairns, B. R., Kim, Y. J., Sayre, M. H., Laurent, B. C. & Kornberg, R. D. A multisubunit complex containing the SWI1/ADR6, SWI2/SNF2, SWI3, SNF5, and SNF6 gene products isolated from yeast. Proc. Natl Acad. Sci. USA 91, 1950–1954 (1994).
Kadoch, C. & Crabtree, G. R. Mammalian SWI/SNF chromatin remodeling complexes and cancer: mechanistic insights gained from human genomics. Sci. Adv. 1, e1500447 (2015).
Sen, P. et al. Loss of Snf5 induces formation of an aberrant SWI/SNF complex. Cell Rep. 18, 2135–2147 (2017).
Beauchamp, R. L. et al. A high-throughput kinome screen reveals serum/glucocorticoid-regulated kinase 1 as a therapeutic target for NF2-deficient meningiomas. Oncotarget 6, 16981–16997 (2015).
Vierbuchen, T. et al. AP-1 transcription factors and the BAF complex mediate signal-dependent enhancer selection. Mol. Cell 68, 1067–1082.e12 (2017).
Pan, J. et al. Interrogation of mammalian protein complex structure, function, and membership using genome-scale fitness screens. Cell Syst. 6, 555–568.e7 (2018).
Gatchalian, J. et al. A non-canonical BRD9-containing BAF chromatin remodeling complex regulates naive pluripotency in mouse embryonic stem cells. Nat. Commun. 9, 5139 (2018).
Wang, X. et al. BRD9 defines a SWI/SNF sub-complex and constitutes a specific vulnerability in malignant rhabdoid tumors. Nat. Commun. 10, 1881 (2019).
Remillard, D. et al. Degradation of the BAF complex factor BRD9 by heterobifunctional ligands. Angew. Chem. Int. Ed. Engl. 56, 5738–5743 (2017).
Brien, G. L. et al. Targeted degradation of BRD9 reverses oncogenic gene expression in synovial sarcoma. eLife 7, e41305 (2018).
Papillon, J. P. N. et al. Discovery of orally active inhibitors of brahma homolog (BRM)/SMARCA2 ATPase activity for the treatment of brahma related gene 1 (BRG1)/SMARCA4-mutant cancers. J. Med. Chem. 61, 10155–10172 (2018).
Kadoch, C. & Crabtree, G. R. Reversible disruption of mSWI/SNF (BAF) complexes by the SS18-SSX oncogenic fusion in synovial sarcoma. Cell 153, 71–85 (2013).
Corces, M. R. et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods 14, 959–962 (2017).
Buenrostro, J. D., Wu, B., Chang, H. Y. & Greenleaf, W. J. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 109, 21.29.1–21.29.9 (2015).
Pettersen, E. F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput Chem. 25, 1605–1612 (2004).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Ramírez, F. et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137–R137 (2008).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Picard. http://broadinstitute.github.io/picard/ or https://github.com/broadinstitute/picard
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Zhu, Q., Liu, N., Orkin, S. H. & Yuan, G.-C. CUT&RUNTools: a flexible pipeline for CUT&RUN processing and footprint analysis. Genome Biol. 20, 192–192 (2019).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Kuhn, R. M., Haussler, D. & Kent, W. J. The UCSC genome browser and associated tools. Brief. Bioinforma. 14, 144–161 (2013).
Subramanian, A. et al. Gene Set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Tripathi, S. et al. Meta- and orthogonal integration of influenza “OMICs” data defines a role for UBR4 in virus budding. Cell Host Microbe 18, 723–735 (2015).
Schäfer, J. & Strimmer, K. A shrinkage approach to large-scale covariance matrix estimation and implications for functional genomics. Stat. Appl. Genet. Mol. Biol. 4, 32 (2005).
Opgen-Rhein, R. & Strimmer, K. Accurate ranking of differentially expressed genes by a distribution-free shrinkage approach. Stat. Appl. Genet. Mol. Biol. 6, 9 (2007).
Shen, L., Shao, N., Liu, X. & Nestler, E. ngs.plot: quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genom. 15, 284–284 (2014).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Sheffield, N. C. & Bock, C. LOLA: enrichment analysis for genomic region sets and regulatory elements in R and Bioconductor. Bioinformatics 32, 587–589 (2016).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B Cell Identities. Mol. Cell 38, 576–589 (2010).
Schaar, B. T. et al. GREAT improves functional interpretation of cis -regulatory regions. Nat. Biotechnol. 28, 495–501 (2010).
Schep, A. N., Wu, B., Buenrostro, J. D. & Greenleaf, W. J. chromVAR: inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nat. Methods 14, 975–978 (2017).
Vierstra, J. et al. Global reference mapping of human transcription factor footprints. Nature 583, 729–736 (2020).
Acknowledgements
We thank all members of the Kadoch laboratory for thoughtful discussions throughout the duration of this project. We are grateful to Z. Herbert and M. Sullivan of the DFCI Molecular Biology Core Facility (MBCF) for help with high-throughput sequencing studies. We are grateful to M. Leidl for technical assistance in SMARCE1 cloning and biochemical experiments. We thank N. Gray for the synthesis and scale-up of dBRD9A. This study was supported in part by the Landry Cancer Consortium Award (R.S.P.), the National Institutes of Health 5F31CA228441-02 (R.S.P.), grants 1DP2CA195762-01 (C.K.), the Pew-Stewart Scholars in Cancer Research Award (C.K.), American Brain Tumor Association (ABTA), the American Cancer Society Research Scholar Award RSG-14-051-01-DMC (C.K.), the all-Manchester National Institute for Health and Care Research (NIHR) Biomedical Research Centre (IS-BRC-1215-20007) (M.J.S.), and the U.S. Army Medical Research Acquisition Activity Congressionally Directed Medical Research Program (USAMRAA CDMRP) Neurofibromatosis Research Program, Investigator-Initiated Research Award (W81XWH1910334) (M.J.S.).
Author information
Authors and Affiliations
Contributions
R.S.P., C.J.W. and C.K. conceived of the study. R.S.P. performed all experiments with help from C.J.W., D.D.S.G., O.B. and N.M. All computational analyses were performed by C.K.C., with help from A.S. for cross-linking mass spectrometry and structural analyses. Cmp12 was synthesized and validated by Y.L. with oversight from J.Q., M.J.S., E.H.G., W.L.B. and D.M.M. identified and collected primary tumor and normal human tissue for DNA and RNA sequencing. V.R. provided the AC7 arachnoid cell line, the AC7 NF2-deleted cell line, and guidance for CRISPR–Cas9-mediated editing experiments. C.K. supervised the study. R.S.P., C.K. and C.K.C. wrote and edited the paper.
Corresponding author
Ethics declarations
Competing interests
C.K. is the scientific founder, Fiduciary Board of Directors member, Scientific Advisory Board member, shareholder and consultant for Foghorn Therapeutics, Inc. (Cambridge, MA). C.K. is also a member of the Scientific Advisory Boards of Nereid Therapeutics and Nested Therapeutics, and serves as a consultant for Cell Signaling Technologies. All other authors do not declare any competing interests.
Peer review
Peer review information
Nature Genetics thanks Blaine Bartholomew, Tom Owen-Hughes and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 CCM-associated loss of SMARCE1 selectively impacts the cBAF assembly of mSWI/SNF complexes.
a. Distribution of cranial and spinal SMARCE1-deficient clear cell meningioma reported in the literature; male and female cases indicated in legend. Image adapted from ‘CNS (lateral, no nerves)’, by BioRender.com (2022). Retrieved from https://app.biorender.com/biorender-templates. b. Age distribution among SMARCE1-deficient CCM cases (n = 71); median= 10.8 years. c. Frequently mutated genes and their associated pathways in meningioma. SMARCE1 loss uniformly defines the clear cell meningioma subtype. d. Domain architecture and conservation of the human SMARCE1 protein. e. Schematic for SMARCE1 rescue experiments in BT549 SMARCE1-deificient cells. f. Density sedimentation experiments using 10–30% glycerol gradients performed on nuclear extracts isolated from BT549 cells infected with either empty vector control or WT SMARCE1. g. Lentiviral expression of SMARCE1 (or GFP control) in BT549 (SMARCE1 -/-) cells, followed by IP-western blotting for mSWI/SNF complex subunits. *IgG h.c. indicates IgG heavy chain bands are present. (f-g), representative of n = 3 experiments with similar results.
Extended Data Fig. 2 SMARCE1 has structural homology to ySnf6 and tethers to the initial core of cBAF and PBAF complexes, but to ARID1A and SMARCA4 only in cBAF complexes.
a,b. CX-MS analyses performed on (A) apo human canonical BAF (cBAF), and (B) NCP-bound cBAF complexes. SMARCE1, cBAF-specific subunits (ARID1A and DPF2), and the ATPase subunit, SMARCA4, are shown. c. CX-MS analyses performed on apo human PBAF complexes. SMARCE1, PBAF-specific subunits (ARID2, PBRM1, BRD7 and PHF10), and the ATPase, SMARCA4, are shown. d. Top, Number and distribution of lysine (K) residues (red) on ARID1A/B and ARID2 subunits. Core binding region (CBR) on each is indicated. Bottom, pairwise alignment between ARID1A/B and ARID2 with SMARCE1-ARID1A CX-MS sites highlighted (purple). e. Cross-linking mass spec analyses performed on ySWI/SNF complexes in nucleosome-unbound states [Sen et al., 2017; Mashtalir et al., 2018]. Crosslinks are plotted as a percentage of total crosslinks recovered in the dataset, and are marked by the ySWI/SNF subunit to which they tether (legend). Selected crosslinked lysine (K) residues of Snf6 are labeled. DeepCoil coiled-coil prediction scores are indicated on top row. f. Amino acid sequence conservation of human SMARCE1 to yeast Snf6. g. Human cBAF (PDB: 6LTJ) structure contrasted to cBAF-like yeast SWI/SNF (PDB: 6UXW). h. Cryo-EM structures of ySWI/SNF (PDB: 6UXW) and yRSC (PDB: 6TDA and PDB: 6KW4). Putative SMARCE1 homologs, Snf6 and Htl1, are highlighted in red.
Extended Data Fig. 3 The SMARCE1 subunit in cBAF complexes is required for cBAF-mediated enhancer accessibility.
a. Top, Metaplots over all peaks from Fig. 3b for SMARCE1, SMARCA4, ARID1A, DPF2, ARID2, SS18, H3K27Ac, H3K4me3, and ATAC-seq. Solid line, wild-type AC7 cells; dashed line, SMARCE1 KO cells; Bottom, metaplots over each cluster (clusters 1–3). b. Correlation plots of RNA-seq data between two independent SMARCE1-KO AC7 clones. c. Proliferation of BT549 cells with GFP control or WT SMARCE1 rescue. d. Immunoblot performed on nuclear protein isolated from BT549 cells rescued with control GFP, and wild type SMARCE1. e. Metaplots for ChIP-seq performed on BT549 cells in naive (GFP control infected) or +SMARCE1 conditions (over all peaks from Fig. 3h). f. Stacked bar graph indicating distribution of shared and gained SMARCC1/SMARCA4 peaks upon SMARCE1 rescue in BT549 cells by distance to TSS. g. MA plot reflecting accessibility changes upon SMARCE1 rescue in BT549 cells. h. Stacked bar graph indicating distribution of shared and gained DNA accessibility (ATAC-seq) peaks upon SMARCE1 rescue in BT549 cells by distance to TSS. i. Venn Diagram reflecting overlap between gained SMARCA4/SMARCC1 merged peaks and gained ATAC-seq peaks in BT549 cells in the + SMARCE1 rescue condition. j. Box and whisker plot reflecting gene expression LogFC across sites indicated (gained BAF complex target sites, gained DNA accessibility sites, and BAF/accessibility dually gained sites in BT549 cells with +SMARCE1 rescue. k. Venn diagram of SMARCA4 sites in SMARCE1-KO AC7 cells rescued with either GFP control or WT SMARCE1. l. Heatmap for all merged SMARCA4 sites depicting SMARCE1, SMARCA4, H3K27Ac ChIP-seq, and ATAC-seq in SMARCE1-KO AC7 cells rescued with either GFP control or WT SMARCE1.
Extended Data Fig. 4 SMARCE1 loss alters transcriptome in SMARCE1-deficient AC7 cells and in clear cell meningioma tumors.
a. MA plot showing gene expression changes upon SMARCE1 loss in AC7 cells. Red and blue dots represent genes significantly upregulated and downregulated, respectively (adj. P < 0.01). b. Motif enrichment analysis by HOMER over sites of BAF occupancy loss in SMARCE1-deficient AC7 cells and of BAF occupancy gain in SMARCE1-deficient cells rescued by WT SMARCE1. c. (Left) Venn diagram reflecting overlap among sites with SMARCA4 loss, sites with accessibility loss, and sites nearest to genes with significant decreases in expression in SMARCE1-deficient AC7 relative to WT (from Cluster 2 in Fig. 3b); (right) Lollipop plot displaying expression of select 55 genes downregulated in AC7 cells that are near sites with decreases in both BAF binding and accessibility. d. Boxplots displaying expression levels of SMARCE1 and NF2 genes in control, SMARCE1-deficient, and NF2-deficient AC7 cells (data from n = 3 biologically-independent cell lines, for normal and SMARCE1-null, and n = 1 cell line for NF2-null; center represents mean, whiskers represent 1.5*IQR, bound of box represent 25th and 75th percentiles for RPKM values). e. Immunoblot performed on nuclear protein isolated from NF2-deficient AC7 clones with associated control; representative of n = 3 experiments with similar results. f. MA plot showing gene expression changes upon NF2 loss in AC7 cells. Red and blue dots represent genes significantly upregulated and downregulated, respectively (adj. P < 0.01). g,h. Gene ontology analysis by Metascape on up and down DEGs (adj. P < 0.01) in SMARCE1-deficient and NF2-deficient AC7 cells relative to the WT condition. i. GSEA results displaying Hallmark MTORC1 Signaling and MYC Targets gene set enrichment for NF2 deficient AC7 cells. j. Volcano plots showing differentially expressed genes in SMARCE1-deficient, NF2-deficient, and KLF/TRAF7-deficient clear cell meningioma tumors relative to normal tissues. Red and blue dots represent genes significantly upregulated and downregulated DEGs, respectively (adj. P < 0.05). k. Venn diagrams reflecting overlap of upregulated and downregulated DEGs for SMARCE1-deficient, NF2-deficient, and KLF/TRAF7-deficient clear cell meningioma tumors relative to normal tissues. l. Heatmap of combined differentially expressed genes from tumor RNA-seq data shown in Fig. 4m. After Z-score transformation, K-means clustering was used to partition the data into 4 groups. m. Gene set enrichment results from hypergeometric tests (using Wikipathway MSIGDB gene set collection) from clusters from Fig. 4g. n. Bar plots displaying gene expression for select genes (FLNA, EBF1, STXBP1, and SKP2) in WT or normal, SMARCE1-deficient, and NF2-deficient AC7 cells and CCM tumors from clusters in Fig. 4g (Mean with error bars representing S.D. are shown for primary tumors, derived from n = />3 biological replicates (samples)).
Extended Data Fig. 5 Increased biochemical assembly and targeting of ncBAF complexes upon loss of the SMARCE1 subunit.
a. Metaplots for SMARCE1, SMARCA4, SS18, BRD9, H3K27Ac, and ATAC-seq performed over Cluster 3 in Fig. 3b. b. ECDF plots of SMARCA4 Log2 fold change (relative to WT) as a function of motif count for 286 non-redundant transcription factor archetypes motifs over merged BAF peaks in AC7 SMARCE1-KO cells rescued with WT SMARCE1. c. Venn diagrams reflecting (top) all Cut&Tag peaks for BRD9 and SMARCD1 in SMARCE1 WT and KO conditions in AC7 cells; (bottom) Venn diagrams reflecting all ChIP-seq peaks for SMARCA4, ARDI1A, DPF2 in SMARCE1 WT and KO conditions in AC7 cells. d. HOMER motif enrichment performed on Cluster 2 (gained) sites in Fig. 5f. e. Distance to TSS stacked bar graphs corresponding to retained, gained, and lost clusters of SMARCD1 and BRD9 sites identified in Fig. 5f. f. Example tracks at the MS4A7 locus. Tracks for mSWI/SNF subunits, H3K27ac mark, as well as ATAC-seq and RNA-seq tracks are shown.
Extended Data Fig. 6 ncBAF inhibition as synthetic lethal strategy in SMARCE1-deficient clear cell meningioma.
a. Distance to TSS stacked bar graphs for differentially accessible sites (adj P < 0.05) following dBRD9A and CMP12 treatment in SMARCE1-KO and WT AC7 cells. b. Motif enrichment analysis by HOMER over sites with accessibility loss in SMARCE1-deficient AC7 cells after dBRD9A treatment (adj P < 0.05). c. Dose response curves showing CMP12 treatments of WT, SMARCE1-KO, and NF2-KO AC-7 cell lines (n = 2; Mean with SD). d. Proliferation curves for WT and NF2-KO clones treated with 50 nM of CMP12 over the days indicated. Error bars represent S.D. of mean, and p-values derived from two-sided t test are shown (n.s., non-significant), data from n = 3 biologically-independent experiments. e. Stacked bar plots displaying numbers of differentially expressed genes after CMP12 treatment in WT and SMARCE1-deficient AC7 cells. f. (Left) Volcano plot showing differentially expressed genes in SMARCE1-deficient AC7 cells after CMP12 treatment relative to DMSO. Blue dots represent genes with significantly downregulated DEGs (adj. P < 0.01) and purple dots represent downregulated genes that map to sites with accessibility loss in the SMARCE1-deficient cells but not WT cells after dBRD9A treatment. (Right) Pie chart characterizes the distribution of downregulated genes in the SMARCE1-deficient cells after dBRD9A treatment. g. PCA Analysis of ATAC-seq data over merged ATAC-seq peaks for dBRD9A (top) and CMP12 (bottom) treatment in WT and SMARCE1-deficient AC7 cells. h, i. Cis-regulatory analysis by GREAT on differentially accessible sites after dBRD9A treatment (top) and CMP12 treatment (bottom) in SMARCE1-deficient AC7 cells using Go Biological Process terms. j. Bar plots displaying numbers of differentially expressed genes after CMP12 treatment in WT and SMARCE1-deficient AC7 cells at three concentrations relative to changes between WT and SMARCE1-deficient cells in the DMSO condition. k. Venn diagrams show the overlap of upregulated and downregulated DEGs between KO and WT conditions at three concentrations relative to the differences between the KO and WT in the DMSO condition. l. GSEA results showing gene set enrichment (using Hallmark MSIGDB gene set collection) for the KO vs. WT in DMSO comparison and across 3 concentrations of CMP12. m. (left) Venn diagram highlighting downregulated DEGs after dBRD9A/CMP12 treatment (union) in SMARCE1-deficient cells that are not downregulated in WT cells. (Right) Gene ontology analysis performed on genes downregulated following dBRD9A/CMP12 treatment (union) in SMARCE1-KO but not WT cells. n. (Left) Venn diagram highlighting upregulated DEGs after dBRD9A/CMP12 treatment (union) in SMARCE1-deficient cells that are not upregulated in WT cells. (Right) Gene ontology analysis performed on DEGs in red shaded region of Venn diagram on left. o. Expression of top most variable genes reversed in expression by both CMP12 and dBRD9A treatments in SMARCE1-KO cells.
Supplementary information
Supplementary Tables
Supplementary Table 1. SMARCE1-mutant clear cell meningoma cases from the literature. Supplementary Table 2. mSWI/SNF cross-linking mass spectrometry data and calculations. Supplementary Table 3. Details of primary meningioma and normal meninges samples used in this study. Supplementary Table 4. Antibody information.
Source data
Source Data Fig. 1
Unprocessed western blots.
Source Data Fig. 2
Unprocessed western blots.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Unprocessed western blots.
Source Data Fig. 6
Unprocessed western blots.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 7
Unprocessed western blots.
Source Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 1
Unprocessed western blots.
Source Data Extended Data Fig. 3
Unprocessed western blots.
Source Data Extended Data Fig. 4
Unprocessed western blots.
Source Data Extended Data Fig. 6
Statistical source data.
Rights and permissions
About this article
Cite this article
St. Pierre, R., Collings, C.K., Samé Guerra, D.D. et al. SMARCE1 deficiency generates a targetable mSWI/SNF dependency in clear cell meningioma. Nat Genet 54, 861–873 (2022). https://doi.org/10.1038/s41588-022-01077-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-022-01077-0
This article is cited by
-
BCL7A and BCL7B potentiate SWI/SNF-complex-mediated chromatin accessibility to regulate gene expression and vegetative phase transition in plants
Nature Communications (2024)
-
DNA methylation provides diagnostic value for meningioma recurrence in clinical practice
Acta Neurochirurgica (2023)
-
The multiomic landscape of meningiomas: a review and update
Journal of Neuro-Oncology (2023)
-
(mis)-Targeting of SWI/SNF complex(es) in cancer
Cancer and Metastasis Reviews (2023)