Abstract
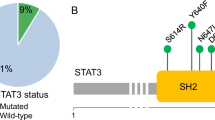
Chronic lymphoproliferative disorder of natural killer cells (CLPD-NK) is characterized by clonal expansion of natural killer (NK) cells where the underlying genetic mechanisms are incompletely understood. In the present study, we report somatic mutations in the chemokine gene CCL22 as the hallmark of a distinct subset of CLPD-NK. CCL22 mutations were enriched at highly conserved residues, mutually exclusive of STAT3 mutations and associated with gene expression programs that resembled normal CD16dim/CD56bright NK cells. Mechanistically, the mutations resulted in ligand-biased chemokine receptor signaling, with decreased internalization of the G-protein-coupled receptor (GPCR) for CCL22, CCR4, via impaired β-arrestin recruitment. This resulted in increased cell chemotaxis in vitro, bidirectional crosstalk with the hematopoietic microenvironment and enhanced NK cell proliferation in vivo in transgenic human IL-15 mice. Somatic CCL22 mutations illustrate a unique mechanism of tumor formation in which gain-of-function chemokine mutations promote tumorigenesis by biased GPCR signaling and dysregulation of microenvironmental crosstalk.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Primary sample whole-genome and transcriptome data have been deposited in the European Genome–phenome Archive (EGA), accession no. EGAS00001006009, and are also housed in a protected Cloud environment in accordance with European General Data Protection regulations. Transcriptome sequencing data (genome build GRCh37) of engrafted NK-92 cells have been deposited in the Gene Expression Omnibus, accession no. GSE163864. Normal NK cell data were downloaded from GSE133383 (ref. 77). These data were used for Figs. 1, 2 and 4, and Extended Data Figs. 1–3 and 10. Source data are provided with this paper.
Code availability
All details of all analysis code used in the manuscript has been provided in Methods.
References
Swerdlow, S. H. et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood 127, 2375–2390 (2016).
Barila, G. et al. T cell large granular lymphocyte leukemia and chronic NK lymphocytosis. Best. Pract. Res. Clin. Haematol. 32, 207–216 (2019).
Lamy, T., Moignet, A. & Loughran, T. P. Jr LGL leukemia: from pathogenesis to treatment. Blood 129, 1082–1094 (2017).
Giussani, E. et al. Lack of viral load within chronic lymphoproliferative disorder of natural killer cells: what is outside the leukemic clone? Front Oncol. 10, 613570 (2020).
Lamy, T. & Loughran, T. P. Jr How I treat LGL leukemia. Blood 117, 2764–2774 (2011).
Morice, W. G. et al. Chronic lymphoproliferative disorder of natural killer cells: a distinct entity with subtypes correlating with normal natural killer cell subsets. Leukemia 24, 881–884 (2010).
Barila, G. et al. Dominant cytotoxic NK cell subset within CLPD-NK patients identifies a more aggressive NK cell proliferation. Blood Cancer J. 8, 51 (2018).
Smith, S. L. et al. Diversity of peripheral blood human NK cells identified by single-cell RNA sequencing. Blood Adv. 4, 1388–1406 (2020).
Wagner, J. A. et al. CD56bright NK cells exhibit potent antitumor responses following IL-15 priming. J. Clin. Invest. 127, 4042–4058 (2017).
Michel, T. et al. Human CD56bright NK cells: an update. J. Immunol. 196, 2923–2931 (2016).
Dogra, P. et al. Tissue determinants of human NK cell development, function, and residence. Cell 180, 749–763.e13 (2020).
Koskela, H. L. et al. Somatic STAT3 mutations in large granular lymphocytic leukemia. N. Engl. J. Med. 366, 1905–1913 (2012).
Jerez, A. et al. STAT3 mutations unify the pathogenesis of chronic lymphoproliferative disorders of NK cells and T-cell large granular lymphocyte leukemia. Blood 120, 3048–3057 (2012).
Gasparini, V. R. et al. A high definition picture of somatic mutations in chronic lymphoproliferative disorder of natural killer cells. Blood Cancer J. 10, 42 (2020).
Pastoret, C. et al. Linking the KIR phenotype with STAT3 and TET2 mutations to identify chronic lymphoproliferative disorders of NK cells. Blood 137, 3237–3250 (2021).
Olson, T. L. et al. Frequent somatic TET2 mutations in chronic NK-LGL leukemia with distinct patterns of cytopenias. Blood 138, 662–673 (2021).
Cheon, H. et al. Analysis of genomic landscape of large granular lymphocyte leukemia reveals etiologic insights. Blood 136, 27–28 (2020).
Stengel, A., Meggendorfer, M., Kern, W., Haferlach, T. & Haferlach, C. Correlation of mutation patterns with patient age in 2656 cases with 11 different hematological malignancies. Blood 136, 16–17 (2020).
Jiang, L. et al. Exome sequencing identifies somatic mutations of DDX3X in natural killer/T-cell lymphoma. Nat. Genet. 47, 1061–1066 (2015).
Dufva, O. et al. Aggressive natural killer-cell leukemia mutational landscape and drug profiling highlight JAK-STAT signaling as therapeutic target. Nat. Commun. 9, 1567 (2018).
Scheu, S., Ali, S., Ruland, C., Arolt, V. & Alferink, J. The C-C chemokines CCL17 and CCL22 and their receptor CCR4 in CNS autoimmunity. Int. J. Mol. Sci. 18, 2306 (2017).
Homey, B., Steinhoff, M., Ruzicka, T. & Leung, D. Y. Cytokines and chemokines orchestrate atopic skin inflammation. J. Allergy Clin. Immunol. 118, 178–189 (2006).
Lucas, M., Schachterle, W., Oberle, K., Aichele, P. & Diefenbach, A. Dendritic cells prime natural killer cells by trans-presenting interleukin 15. Immunity 26, 503–517 (2007).
Anguille, S. et al. Interleukin-15 dendritic cells harness NK cell cytotoxic effector function in a contact- and IL-15-dependent manner. PLoS ONE 10, e0123340 (2015).
Andoniou, C. E. et al. Interaction between conventional dendritic cells and natural killer cells is integral to the activation of effective antiviral immunity. Nat. Immunol. 6, 1011–1019 (2005).
Zambello, R. et al. Phenotypic and functional analyses of dendritic cells in patients with lymphoproliferative disease of granular lymphocytes (LDGL). Blood 106, 3926–3931 (2005).
Godiska, R. et al. Human macrophage–derived chemokine (MDC), a novel chemoattractant for monocytes, monocyte-derived dendritic cells, and natural killer cells. J. Exp. Med. 185, 1595–1604 (1997).
Zheng, Y. et al. Structure of CC chemokine receptor 5 with a potent chemokine antagonist reveals mechanisms of chemokine recognition and molecular mimicry by HIV. Immunity 46, 1005–1017.e5 (2017).
Salanga, C. L. & Handel, T. M. Chemokine oligomerization and interactions with receptors and glycosaminoglycans: the role of structural dynamics in function. Exp. Cell. Res. 317, 590–601 (2011).
Wacker, D. et al. Crystal structure of an LSD-bound human serotonin receptor. Cell 168, 377–389.e12 (2017).
McCorvy, J. D. et al. Structural determinants of 5-HT2B receptor activation and biased agonism. Nat. Struct. Mol. Biol. 25, 787–796 (2018).
Yamashita, U. & Kuroda, E. Regulation of macrophage-derived chemokine (MDC, CCL22) production. Crit. Rev. Immunol. 22, 105–114 (2002).
Yoshie, O. & Matsushima, K. CCR4 and its ligands: from bench to bedside. Int. Immunol. 27, 11–20 (2014).
Mariani, M., Lang, R., Binda, E., Panina-Bordignon, P. & D’Ambrosio, D. Dominance of CCL22 over CCL17 in induction of chemokine receptor CCR4 desensitization and internalization on human Th2 cells. Eur. J. Immunol. 34, 231–240 (2004).
Rapp, M. et al. CCL22 controls immunity by promoting regulatory T cell communication with dendritic cells in lymph nodes. J. Exp. Med. 216, 1170–1181 (2019).
Curiel, T. J. et al. Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat. Med. 10, 942–949 (2004).
Mailloux, A. W. & Young, M. R. I. NK-dependent Increases in CCL22 secretion selectively recruits regulatory T cells to the tumor microenvironment. J. Immunol. 182, 2753–2765 (2009).
Nakagawa, M. et al. Gain-of-function CCR4 mutations in adult T cell leukemia/lymphoma. J. Exp. Med. 211, 2497–2505 (2014).
Smith, J. S., Lefkowitz, R. J. & Rajagopal, S. Biased signalling: from simple switches to allosteric microprocessors. Nat. Rev. Drug Discov. 17, 243–260 (2018).
Lefkowitz, R. J. & Shenoy, S. K. Transduction of receptor signals by beta-arrestins. Science 308, 512–517 (2005).
Drury, L. J. et al. Monomeric and dimeric CXCL12 inhibit metastasis through distinct CXCR4 interactions and signaling pathways. Proc. Natl Acad. Sci. USA 108, 17655–17660 (2011).
Bernardini, G., Sciume, G. & Santoni, A. Differential chemotactic receptor requirements for NK cell subset trafficking into bone marrow. Front. Immunol. 4, 12 (2013).
Wright, D. E., Bowman, E. P., Wagers, A. J., Butcher, E. C. & Weissman, I. L. Hematopoietic stem cells are uniquely selective in their migratory response to chemokines. J. Exp. Med. 195, 1145–1154 (2002).
Eisenman, J. et al. Interleukin-15 interactions with interleukin-15 receptor complexes: characterization and species specificity. Cytokine 20, 121–129 (2002).
Costantini, C. et al. Neutrophil activation and survival are modulated by interaction with NK cells. Int. Immunol. 22, 827–838 (2010).
Thoren, F. B. et al. Human NK cells induce neutrophil apoptosis via an NKp46- and Fas-dependent mechanism. J. Immunol. 188, 1668–1674 (2012).
Park, C. S., Yoon, S. O., Armitage, R. J. & Choi, Y. S. Follicular dendritic cells produce IL-15 that enhances germinal center B cell proliferation in membrane-bound form. J. Immunol. 173, 6676–6683 (2004).
Katakai, T. Marginal reticular cells: a stromal subset directly descended from the lymphoid tissue organizer. Front. Immunol. 3, 200 (2012).
Li, L., Wu, J., Abdi, R., Jewell, C. M. & Bromberg, J. S. Lymph node fibroblastic reticular cells steer immune responses. Trends Immunol. 42, 723–734 (2021).
The ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium. Pan-cancer analysis of whole genomes. Nature 578, 82–93 (2020).
Wang, Z. Q. et al. Gain-of-function mutation of KIT ligand on melanin synthesis causes familial progressive hyperpigmentation. Am. J. Hum. Genet 84, 672–677 (2009).
Wootten, D., Christopoulos, A., Marti-Solano, M., Babu, M. M. & Sexton, P. M. Mechanisms of signalling and biased agonism in G protein-coupled receptors. Nat. Rev. Mol. Cell Biol. 19, 638–653 (2018).
Caligiuri, M. A. Human natural killer cells. Blood 112, 461–469 (2008).
Ali, A. K., Nandagopal, N. & Lee, S. H. IL-15-PI3K-AKT-mTOR: a critical pathway in the life journey of natural killer cells. Front. Immunol. 6, 355 (2015).
Mishra, A. et al. Aberrant overexpression of IL-15 initiates large granular lymphocyte leukemia through chromosomal instability and DNA hypermethylation. Cancer Cell 22, 645–655 (2012).
Ferlazzo, G. & Morandi, B. Cross-talks between natural killer cells and distinct subsets of dendritic cells. Front. Immunol. 5, 159 (2014).
Liu, B. et al. Affinity-coupled CCL22 promotes positive selection in germinal centres. Nature 592, 133–137 (2021).
Bakker, E., Qattan, M., Mutti, L., Demonacos, C. & Krstic-Demonacos, M. The role of microenvironment and immunity in drug response in leukemia. Biochim. Biophys. Acta 1863, 414–426 (2016).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Ajram, L. et al. Internalization of the chemokine receptor CCR4 can be evoked by orthosteric and allosteric receptor antagonists. Eur. J. Pharmacol. 729, 75–85 (2014).
Mortier, E., Woo, T., Advincula, R., Gozalo, S. & Ma, A. IL-15Ralpha chaperones IL-15 to stable dendritic cell membrane complexes that activate NK cells via trans presentation. J. Exp. Med. 205, 1213–1225 (2008).
Nandagopal, N., Ali, A. K., Komal, A. K. & Lee, S. H. The critical role of IL-15–PI3K–mTOR pathway in natural killer cell effector functions. Front. Immunol. 5, 187 (2014).
Crinier, A., Narni-Mancinelli, E., Ugolini, S. & Vivier, E. SnapShot: natural killer cells. Cell 180, 1280–1280.e1 (2020).
Freud, A. G., Mundy-Bosse, B. L., Yu, J. & Caligiuri, M. A. The broad spectrum of human natural killer cell diversity. Immunity 47, 820–833 (2017).
Geiger, T. L. & Sun, J. C. Development and maturation of natural killer cells. Curr. Opin. Immunol. 39, 82–89 (2016).
Gaggero, S., Witt, K., Carlsten, M. & Mitra, S. Cytokines orchestrating the natural killer–myeloid cell crosstalk in the tumor microenvironment: implications for natural killer cell-based cancer immunotherapy. Front. Immunol. 11, 621225 (2020).
Michel, T., Hentges, F. & Zimmer, J. Consequences of the crosstalk between monocytes/macrophages and natural killer cells. Front. Immunol. 3, 403 (2012).
Rohrle, N., Knott, M. M. L. & Anz, D. CCL22 signaling in the tumor environment. Adv. Exp. Med. Biol. 1231, 79–96 (2020).
Fehniger, T. A. et al. Fatal leukemia in interleukin 15 transgenic mice follows early expansions in natural killer and memory phenotype CD8+ T cells. J. Exp. Med. 193, 219–231 (2001).
Raczy, C. et al. Isaac: ultra-fast whole-genome secondary analysis on Illumina sequencing platforms. Bioinformatics 29, 2041–2043 (2013).
Kim, S. et al. Strelka2: fast and accurate calling of germline and somatic variants. Nat. Methods 15, 591–594 (2018).
Karczewski, K. J. et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443 (2020).
Haferlach, T., Hutter, S. & Meggendorfer, M. Genome sequencing in myeloid cancers. N. Engl. J. Med. 384, e106 (2021).
Hutter, S. et al. A novel machine learning based in silico pathogenicity predictor for missense variants in a hematological setting Blood 134, 2090 (2019).
Parida, L. et al. Dark-matter matters: discriminating subtle blood cancers using the darkest DNA. PLoS Comput. Biol. 15, e1007332 (2019).
Zhou, X. et al. Exploration of coding and non-coding variants in cancer using GenomePaint. Cancer Cell 39, 83–95.e4 (2021).
Dogra, P. et al. Tissue determinants of human NK. Cell Dev. Funct. Resid. Cell 180, 749–763.e13 (2020).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Anders, S., Pyl, P. T. & Huber, W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Leek, J. T., Johnson, W. E., Parker, H. S., Jaffe, A. E. & Storey, J. D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28, 882–883 (2012).
Zhou, Y. et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523 (2019).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Konagurthu, A. S., Whisstock, J. C., Stuckey, P. J. & Lesk, A. M. MUSTANG: a multiple structural alignment algorithm. Proteins 64, 559–574 (2006).
Sievers, F. et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011).
Valdar, W. S. Scoring residue conservation. Proteins 48, 227–241 (2002).
Wagih, O. ggseqlogo: a versatile R package for drawing sequence logos. Bioinformatics 33, 3645–3647 (2017).
Liu, D., Paczkowski, P., Mackay, S., Ng, C. & Zhou, J. Single-cell multiplexed proteomics on the IsoLight resolves cellular functional heterogeneity to reveal clinical responses of cancer patients to immunotherapies. Methods Mol. Biol. 2055, 413–431 (2020).
Shultz, L. D. et al. Human lymphoid and myeloid cell development in NOD/LtSz-scid IL2R gamma null mice engrafted with mobilized human hemopoietic stem cells. J. Immunol. 174, 6477–6489 (2005).
Aickin, M. & Gensler, H. Adjusting for multiple testing when reporting research results: the Bonferroni vs Holm methods. Am. J. Public Health 86, 726–728 (1996).
Acknowledgements
We thank the American and Lebanese Syrian Associated Charities of St. Jude Children’s Research Hospital and the National Cancer Institute of the NIH for their support under award nos. P30 CA021765, R35 CA197695 (to C.G.M.), R01 CA178393 and P30 CA044579 (to T.P.L.), UG1CA189859, U10CA180820 and UG1CA232760 (to J.F. and E.P.) and F30CA196040 (to A.B.K.); the National Institute of General Medical Sciences grant no. T32 GM080202 (to A.B.K.); the National Library of Medicine grant no. T32 LM012416 (to H.C.); the Henry Schueler 41 & 9 Foundation (to C.G.M.); a St. Baldrick’s Foundation Robert J. Arceci Innovation Award (to C.G.M.); a Garwood Postdoctoral Fellowship of the Hematological Malignancies Program of the St. Jude Children’s Research Hospital Comprehensive Cancer Center (to S.K.); and the Bess Family Charitable Fund, the LGL Leukemia Foundation and a generous anonymous donor (to T.P.L.). We thank ECOG-ACRIN Cancer Research Group for acute NK leukemia samples. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. We thank: S. Surman (Department of Infectious Diseases, St. Jude) for flow cytometry; B. Shemo for LGL Leukemia Registry support; M. Schmachtenberg for technical support; and S. Onengut-Gumuscu and E. Farber for targeted sequencing of the validation cohort at the University of Virginia.
Author information
Authors and Affiliations
Contributions
C.B., S.K., T.L.O., A.R., W.W., C.Q., M.M., M.L.M., C.H., W.K. and I.I. generated genomic data. C.B., S.K., D.J.F., T.L.O., H.C., K.C.O., A.R., J.M.F., T.P.L. and E.P. provided or analyzed patient samples and data. C.B., S.K., W.W. and C.Q. analyzed genomic data. A.K., R.K. and M.M.B. analyzed protein data. S.K., M.S.R., T.F., P.C., S.N.P., S.M.P., P.G.T., L.J. and I.I. conducted experiments. C.B., S.K., I.I., T.H. and C.G.M. designed the study, oversaw the experiments and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
C.B., W.W., M.M. and M.L.M. are employed by MLL Munich Leukemia Laboratory. C.H., W.K. and T.H. have equity ownership of MLL Munich Leukemia Laboratory. I.I. received honoraria from Amgen and Mission Bio. C.G.M. received research funding from Loxo Oncology, Pfizer and AbbVie, and honoraria from Amgen and Illumina, and holds stock in Amgen. T.P.L. is on the Scientific Advisory Board and has stock options for Keystone Nano, Bioniz Therapeutics and Dren Bio. T.P.L. and D.J.F. received honoraria from Kymera Therapeutics. D.J.F. has research funding from AstraZeneca. There are no conflicts of interest with the work presented in this manuscript. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Genetics thanks Gerry Graham, Nicholas Huntington, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 The mutational landscape and clinical parameters CLPD-NK patients.
a, Heatmap of the 59 cases with CLPD-NK in the discovery cohort and 62 CLPD-NK cases in the validation cohort. CCL22 mutations were detected in 16 out of 59 (27%) and 10 out of 62 (16%) cases and were mutually exclusive of STAT3 mutations. Higher CD56 expression was observed in CCL22 mutant cases. b,c, Distribution of CD56 and CD16 expression in the discovery cohort. Hb, hemoglobin; Plt, platelet; WBC, white blood cell; yr, year.
Extended Data Fig. 2 Gene expression profiling of 59 CLPD-NK cases and 12 normal NK samples (6 CD56bright, 6 CD56dim).
A total of 495 differentially expressed genes between CCL22 mutant vs other cases, STAT3 mutant vs other cases, and non-CCL22/STAT3 mutant vs other cases were used for the hierarchical clustering. CCL22 mutant cases were clustered with CD56bright normal NK cells.
Extended Data Fig. 3 CCL22-Pro79Arg transduced NK-92 recapitulated primary CLPD-NK samples with CCL22 mutations in the expression of several important genes.
a, Gene expression ratio of CCL22-Pro79Arg NK-92 cells compared to CCL22-wild type (wt) and empty vector (EV)- transduced NK-92 cells. b, Reduced FASLG expression of CCL22-Pro79Arg NK-92 cells compared to CCL22wt and EV NK-92 cells was confirmed by flow cytometry. CCL22-Pro79Arg NK-92 cells without staining is shown as Negative. c, Immunoblotting confirmed higher GZMK expression in engrafted CCL22-Pro79Arg NK-92 cells. The samples derive from the same experiment and were processed in parallel.
Extended Data Fig. 4 Comparison of growth advantage in CCL22-Pro79arg, wild type, and empty vector transduced cells.
Cytokine independent assay in CCR4 expressing (a) Ba/F3 cells with murine Il-3 or recombinant CCL22 proteins and (b) NK-92 cells with human IL-2, co-expressing wild type or mutant CCL22 (Pro79Arg, Leu45Arg), or GFP-expressing lentiviral vector (Empty vector). No cytokine independent growth was observed. The mean (± SD) cell count (x 1,000,000) is shown (n = 3). c,d, No growth advantage was observed by adding hIL-15 and/or hIL-18 to engineered NK-92 cells. The mean (± SD) cell count (x 1,000,000) is shown (n = 3). e, Recombinant murine IL-15 showed cross-reactivity to human engineered NK-92 cells with lower efficiency than human IL-15. The mean (± SD) relative proliferation rate of murine IL-15 compared to human IL-15 is shown (n = 6, except for empty vector at day6 n = 4 due to technical issue).
Extended Data Fig. 5 Inferring likely functional impact of the mutations on CCL22-CCR4 interactions.
a, 6 missense mutations were identified in CCL22 at 3 positions. The positions at which each mutation occurs are indicated with numbers 1-3, and residue identities for both WT and mutated residues are indicated. Basic residues are highlighted with blue. Mutations of Leu45Arg, Pro46Arg, and Pro79Arg were substitutions to basic residues, suggesting that additional basic residues might modulate chemokine-glycosaminoglycans interactions. b,c, Sequence logo depicting residue conservation among human chemokine paralogs at the five mutated positions. Pro46 and Pro79 were well conserved among all human chemokine paralogs. Structural context of these two conserved Proline residues relative to the chemokine secondary structure: both Prolines occur in loops adjacent to alpha helices and likely facilitate loop turns that orient the adjacent alpha helices. d, Chemokine-GPCR model of the CCL5-CCR5 complex depicting a sulfated Tyrosine in the chemokine receptor N-terminus. Tyrosine sulfation is a common modification of chemokine receptor N-termini with significant effects on chemokine-GPCR interactions and functional properties. e, CCR4 N-terminus with putative sulfotyrosine motif labeled. Chemokine receptors are known to undergo sulfation at tyrosine residues that are adjacent to acidic residues in chemokine receptor N-termini, and sulfated tyrosine residues enhance chemokine-GPCR interactions. f, Comparison of CCL20-CCR6 and CCL5[5P7]-CCR5 chemokine-GPCR complexes reveals that the residue corresponding to CCL22 P79 interacts with the receptor ECL2 in the CCL5[5P7]-CCR5 complex but not the CCL20-CCR6 complex due to different orientations of the chemokine and receptor relative to one another (arrows). g,h, Mutation of the CCL22 residue at P79 may alter the ability of CCL22 to interact with CCR4 at extracellular loop 2. The CCR4 extracellular loop 2 is rich in hydroxyl-containing residues, such that Pro79Arg mutation might be expected to enhance interactions with CCR4 extracellular loop 2 whereas Pro79Leu or Pro79Phe mutations might have minimal or inhibitory effects on CCL22-CCR4 interactions relative to the endogenous Pro79.
Extended Data Fig. 6 Internalization assay with exogenous recombinant wild type or mutant CCL22 in Ba/F3−CCR4 cells.
Both exogenous recombinant wild type and mutant CCL22 internalize CCR4 dose dependently. Mutant CCL22 showed weaker effect than wild type CCL22 in all the tested condition. The mean (± SD) difference from positive control (Ba/F3-CCR4 without treatment; 0%) and negative control (Ba/F3 and Ba/F3-CCR4 with isotype; 100%) is shown (n = 3). P value was calculated by t-test. *0.01 < p < 0.05; **0.001 < p < 0.01.
Extended Data Fig. 7 Paracrine mechanism with endogenous CCL22 induced internalization of CCR4.
a, Immunofluorescence showing (scale bar 10 μm) the difference of CCR4 internalization between CCL22-Pro79Arg and wild type in unpermeabilized and permeabilized condition. CCR4-expressing 293 T cells were transiently transfected with the vector containing CCL22 wild type or Pro79Arg. Unpermeabilized condition only detect surface CCR4, while permeabilized condition can detect both surface and internalized CCR4. Surface CCR4 expression was decreased in wild type CCL22 but not in Pro79Arg-CCL22 mutation after 48 hours incubation. b, Serum swapping assay showing change of CCR4 expression by replacing serum of CCL22 wild type and mutant (Pro79Arg, Leu45Arg) expressing Ba/F3-CCR4 cells. Decreased CCR4 internalization (increased CCR4 expression) was observed by replacing serum of mutant CCL22 from that of CCL22 wild type, and vice versa. The mean (± SD) change rate from serum non-swapped data is shown (n = 3).
Extended Data Fig. 8 CCL22-CCR4 downstream signaling.
a, Phosphorylation of AKT, and mitogen-activated protein kinase (MAPK) (p44/42 extracellular signal-regulated kinase (Erk) 1/2) by immunoblotting. Ba/F3-CCR4 cells were stimulated with exogenous recombinant wild type or mutant CCL22 (50 ng/ml) for 5 minutes (min). Both exogenous recombinant wild type and mutant CCL22 activated pAKT in a similar way, but p42/44 ERK was slightly reduced by mutant CCL22. The samples derive from the same experiment and were processed in parallel. b, Ba/F3-CCR4 cells were stimulated over time with exogenous recombinant wild type or mutant CCL22 (100 ng/ml) and pAKT was assessed by phosphoflow cytometric analysis. Activation of pAKT was not different between exogenous recombinant wild type and mutant CCL22. The mean (± SD) fluorescence intensity (MFI) is shown (n = 3). c,d, Single cell phosphoproteomic detection via IsoLight validated reduced phosphorylation of ERK in mutant CCL22 (50 ng/ml) after three-hour (h) incubation. c, Positive cell rate of pERK and (d) the mean of detected pERK signal are shown. P value was calculated by t-test. **0.001 < p < 0.01.
Extended Data Fig. 9 CCL22 mutations drive proliferation in vivo.
a,b, GFP positive cell rate in (a) spleen and (b) bone marrow cells at the end of study (batch 1; day 57, batch 2; day 44) in NSG mice (3 mice for each group) and NSG-Tg(Hu-IL15) mice that constitutively express human IL-15. Mice were transplanted with NK-92 cells expressing wild type (n = 3 (batch 1, square), 2 (batch 2, round), biologically independent animals) or mutant CCL22 (Pro79Arg; n = 3, 4), or GFP-expressing lentiviral vector (empty vector; n = 3, 4). NK-92 cells expressing mutant CCL22 showed higher GFP positive cell rate than wild type CCL22 and empty vector expressing NK-92 cells in both spleen and bone marrow. GFP positive cells were observed in NSG-Tg(Hu-IL15) mice but not or few in regular NSG mice. The mean GFP positive rate is shown by the dotted line. c,d, Peripheral blood analysis from retroorbital bleeds detected only few circulating human cells at week 6 (batch 2 day 44; the same day of the end of study, empty vector n = 4, wild type CCL22 n = 2, mutant CCL22 n = 4, biologically independent animal samples). Higher circulating human cells were observed in mutant CCL22 although at lower percentage than those observed in spleen and bone marrow. The mean (± SD) GFP positive rate is shown. P value was calculated by t-test. *0.01 < p < 0.05; **0.001 < p < 0.01; ***0.0001 < p < 0.001.
Extended Data Fig. 10 Correction by genome editing of mutated CCL22 reverts the higher expression of chemokine/cytokine and their receptors in primary CLPD-NK samples.
NK cell line NKL which has the hotspot CCL22 mutation (Arg44_Leu45insSer) was edited (CRISPR/Cas9) to revert the mutation (mt) into the wild type (wt) codon. a, Relative gene expression of chemokine receptors in CCL22 mutant compared to wild type was shown. CCL22 mutant NKL represented higher expression of several chemokine receptors. b, Comparison of chemokine and cytokine secretion between CCL22 mutant (NKL-parent-CCL22mt) and wild type NKL (NKL-edited-CCL22wt) by using chemokine array. Cell culture media after 72-hour incubation was analyzed. Several cytokines showed enhanced secretion in CCL22 mutant that related to dendritic cell maturation.
Supplementary information
Supplementary Information
Supplementary Figs. 1-3, Methods and Table information.
Supplementary Tables
Table 1: Patients’ data in discovery and validation cohort. Table 2: Summary of clinical features of CLPD-NK cases with hotspot CCL22 mutations (at residues Leu45 and Pro79). Table 3: The z-score of 59 cases with CLPD-NK and 12 normal NK cells from peripheral blood and BM (acccession no. GSE133383)77. This table describes normalized gene expression of each sample. Table 4: Differentially expressed genes analyzed by DESeq2 between CCL22 mutations versus others, STAT3 mutations versus others and non-CCL22/STAT3 mutations (others) versus others. Table 5: Differentially expressed genes analyzed by DESeq2 between sorted BM/PB CD56bright and CD56dim normal NK cells from the public database (acccession no. GSE133383)77. Table 6: Pathway analysis using differentially expressed genes in CLPD-NK cases with CCL22 mutations, STAT3 mutations and other (non-CCL22/STAT3 mutations), compared with normal NK cell. Table 7: GSEA of 59 cases of primary CLPD-NK patients with CCL22 mutations versus others (c2all). Table 8: The results of intracellular phosphorylation of pERK in BaF3–CCR4 cells after treatment of CCL22 wild-type/mutant proteins detected by isoplexis. Table 9: Differentially expressed genes of engrafted NK-92 cells in NSG-Tg(Hu-IL15) mice analyzed by DESeq2 between CCL22 mutant and wild-type/empty vector-transduced NK-92 cells. Table 10: GSEA of engrafted NK-92 cells in NSG-Tg(Hu-IL15) mice between mutant and wild-type/empty vector-transduced NK-92 cells. Table 11: Gene sets generated by differentially expressed genes for GSEA. Table 12: CCL22-dependent chemokine secretion. This table describes the results from chemokine array performed to compare chemokine secretion between gene-edited CCL22 wild-type and mutant NKL cell lines. Table 13: Antibody list used in the manuscript. Table 14: Primer sequences for cloning and gene editing.
Source data
Source Data Fig. 2
Uncropped western blots for Fig. 2d: actin, GZMK and pSTAT3.
Source Data Extended Data Fig. 3
Uncropped western blots for Fig. 3: GZMK and actin.
Source Data Extended Data Fig. 8
Uncropped western blots for Fig. 8: actin, pAKT and pERK.
Rights and permissions
About this article
Cite this article
Baer, C., Kimura, S., Rana, M.S. et al. CCL22 mutations drive natural killer cell lymphoproliferative disease by deregulating microenvironmental crosstalk. Nat Genet 54, 637–648 (2022). https://doi.org/10.1038/s41588-022-01059-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-022-01059-2
This article is cited by
-
The emerging scenario of immunotherapy for T-cell Acute Lymphoblastic Leukemia: advances, challenges and future perspectives
Experimental Hematology & Oncology (2023)