Abstract
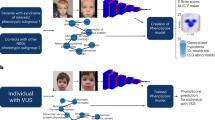
Many monogenic disorders cause a characteristic facial morphology. Artificial intelligence can support physicians in recognizing these patterns by associating facial phenotypes with the underlying syndrome through training on thousands of patient photographs. However, this ‘supervised’ approach means that diagnoses are only possible if the disorder was part of the training set. To improve recognition of ultra-rare disorders, we developed GestaltMatcher, an encoder for portraits that is based on a deep convolutional neural network. Photographs of 17,560 patients with 1,115 rare disorders were used to define a Clinical Face Phenotype Space, in which distances between cases define syndromic similarity. Here we show that patients can be matched to others with the same molecular diagnosis even when the disorder was not included in the training set. Together with mutation data, GestaltMatcher could not only accelerate the clinical diagnosis of patients with ultra-rare disorders and facial dysmorphism but also enable the delineation of new phenotypes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The data that support the findings of this study are divided into two groups, nonsharable data (F2G) and sharable data (OMIM, CASIA-WebFace, GMDB). F2G data are from Face2Gene users and cannot be shared to protect patient privacy. OMIM data can be downloaded at https://omim.org/downloads. CASIA-WebFace and GMDB are available for noncommercial, research and educational purposes, and subject to controlled access. For CASIA-WebFace, user conditions are available at http://www.cbsr.ia.ac.cn/english/casia-webFace/casia-webfAce_AgreEmeNtS.pdf, and requests should be sent to cbsr-request@authenmetric.com. For GMDB, please contact info@gestaltmatcher.org and specify which analyses you intend to perform. The board of GestaltMatcher will check and respond within 10 business days whether your request is compatible with the user conditions.
Code availability
GestaltMatcher can be subdivided into its algorithmic part, data that are required to train the neural network and a service that can be used for matching patients. The project’s landing page, www.gestaltmatcher.org, redirects to separate pages for each category. The web service for matching patients is based on Enc-F2G and is accessible for health care professionals. Parts of this service are proprietary and cannot be shared. However, the architecture of the CNN, as well as the code for evaluation, is available under a creative commons license.
References
Ferreira, C. R. The burden of rare diseases. Am. J. Med. Genet. A 179, 885–892 (2019).
Baird, P. A., Anderson, T. W., Newcombe, H. B. & Lowry, R. B. Genetic disorders in children and young adults: a population study. Am. J. Hum. Genet. 42, 677–693 (1988).
Hart, T. C. & Hart, P. S. Genetic studies of craniofacial anomalies: clinical implications and applications. Orthod. Craniofac. Res. 12, 212–220 (2009).
Marbach, F. et al. The discovery of a LEMD2-associated nuclear envelopathy with early progeroid appearance suggests advanced applications for AI-driven facial phenotyping. Am. J. Hum. Genet. 104, 749–757 (2019).
Ferry, Q. et al. Diagnostically relevant facial gestalt information from ordinary photos. eLife 3, e02020 (2014).
Kuru, K., Niranjan, M., Tunca, Y., Osvank, E. & Azim, T. Biomedical visual data analysis to build an intelligent diagnostic decision support system in medical genetics. Artif. Intell. Med. 62, 105–118 (2014).
Cerrolaza, J. J. et al. Identification of dysmorphic syndromes using landmark-specific local texture descriptors. In 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI) 1080–1083 (IEEE, 2016).
Wang, K. & Luo, J. Detecting visually observable disease symptoms from faces. EURASIP J. Bioinform. Syst. Biol. 2016, 13 (2016).
Dudding-Byth, T. et al. Computer face-matching technology using two-dimensional photographs accurately matches the facial gestalt of unrelated individuals with the same syndromic form of intellectual disability. BMC Biotechnol. 17, 90 (2017).
Shukla, P., Gupta, T., Saini, A., Singh, P. & Balasubramanian, R. A deep learning frame-work for recognizing developmental disorders. In 2017 IEEE Winter Conference on Applications of Computer Vision (WACV) 705–714 (IEEE, 2017).
Liehr, T. et al. Next generation phenotyping in Emanuel and Pallister–Killian syndrome using computer-aided facial dysmorphology analysis of 2D photos. Clin. Genet. 93, 378–381 (2018).
Gurovich, Y. et al. Identifying facial phenotypes of genetic disorders using deep learning. Nat. Med. 25, 60–64 (2019).
van der Donk, R. et al. Next-generation phenotyping using computer vision algorithms in rare genomic neurodevelopmental disorders. Genet. Med. 21, 1719–1725 (2019).
Taigman, Y., Yang, M., Ranzato, M. & Wolf, L. DeepFace: closing the gap to human-level performance in face verification. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition 1701–1708 (IEEE Computer Society, 2014).
Huang, G. B., Ramesh, M., Berg, T. & Learned-Miller, E. Labeled Faces in the Wild: A Database for Studying Face Recognition in Unconstrained Environments. University of Massachusetts, Amherst, Technical Report 07–49 (2007).
Pantel, J. T. et al. Efficiency of computer-aided facial phenotyping (DeepGestalt) in individuals with and without a genetic syndrome: diagnostic accuracy study. J. Med. Internet Res. 22, e19263 (2020).
Landrum, M. J. et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 46, D1062–D1067 (2018).
McKusick, V. A. On lumpers and splitters, or the nosology of genetic disease. Perspect. Biol. Med. 12, 298–312 (1969).
Yi, D., Lei, Z., Liao, S. & Li, S. Z. Learning face representation from scratch. Preprint at arXiv [cs.CV], http://arxiv.org/abs/1411.7923 (2014).
Winter, R. M. & Baraitser, M. The London Dysmorphology Database. J. Med. Genet. 24, 509–510 (1987).
Sobreira, N., Schiettecatte, F., Valle, D. & Hamosh, A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 36, 928–930 (2015).
Stankiewicz, P. et al. Haploinsufficiency of the chromatin remodeler BPTF causes syndromic developmental and speech delay, postnatal microcephaly, and dysmorphic features. Am. J. Hum. Genet. 101, 503–515 (2017).
Morimoto, M. et al. Bi-allelic CCDC47 variants cause a disorder characterized by woolly hair, liver dysfunction, dysmorphic features, and global developmental delay. Am. J. Hum. Genet. 103, 794–807 (2018).
Tanaka, A. J. et al. De novo pathogenic variants in CHAMP1 are associated with global developmental delay, intellectual disability, and dysmorphic facial features. Cold Spring Harb. Mol. Case Stud. 2, a000661 (2016).
Weiss, K. et al. De novo mutations in CHD4, an ATP-dependent chromatin remodeler gene, cause an intellectual disability syndrome with distinctive dysmorphisms. Am. J. Hum. Genet. 99, 934–941 (2016).
Balak, C. et al. Rare de novo missense variants in RNA helicase DDX6 cause intellectual disability and dysmorphic features and lead to P-body defects and RNA dysregulation. Am. J. Hum. Genet. 105, 509–525 (2019).
Harms, F. L. et al. Mutations in EBF3 disturb transcriptional profiles and cause intellectual disability, ataxia, and facial dysmorphism. Am. J. Hum. Genet. 100, 117–127 (2017).
Jansen, S. et al. De novo variants in FBXO11 cause a syndromic form of intellectual disability with behavioral problems and dysmorphisms. Eur. J. Hum. Genet. 27, 738–746 (2019).
Au, P. Y. B. et al. GeneMatcher aids in the identification of a new malformation syndrome with intellectual disability, unique facial dysmorphisms, and skeletal and connective tissue abnormalities caused by de novo variants in HNRNPK. Hum. Mutat. 36, 1009–1014 (2015).
Diets, I. J. et al. De novo and inherited pathogenic variants in KDM3B cause intellectual disability, short stature, and facial dysmorphism. Am. J. Hum. Genet. 104, 758–766 (2019).
Santiago-Sim, T. et al. Biallelic variants in OTUD6B cause an intellectual disability syndrome associated with seizures and dysmorphic features. Am. J. Hum. Genet. 100, 676–688 (2017).
Olson, H. E. et al. A recurrent de novo PACS2 heterozygous missense variant causes neonatal-onset developmental epileptic encephalopathy, facial dysmorphism, and cerebellar dysgenesis. Am. J. Hum. Genet. 102, 995–1007 (2018).
Stephen, J. et al. Bi-allelic TMEM94 truncating variants are associated with neurodevelopmental delay, congenital heart defects, and distinct facial dysmorphism. Am. J. Hum. Genet. 103, 948–967 (2018).
Kanca, O. et al. De novo variants in WDR37 are associated with epilepsy, colobomas, dysmorphism, developmental delay, intellectual disability, and cerebellar hypoplasia. Am. J. Hum. Genet. 105, 413–424 (2019).
Stevens, S. J. C. et al. Truncating de novo mutations in the Krüppel-type zinc-finger gene ZNF148 in patients with corpus callosum defects, developmental delay, short stature, and dysmorphisms. Genome Med. 8, 131 (2016).
Alvi, M., Zisserman, A. & Nellåker, C. Turning a blind eye: explicit removal of biases and variation from deep neural network embeddings. In Computer Vision – ECCV 2018 Workshops 556–572 (Springer International Publishing, 2019).
Lumaka, A. et al. Facial dysmorphism is influenced by ethnic background of the patient and of the evaluator. Clin. Genet. 92, 166–171 (2017).
Schuurs-Hoeijmakers, J. H. M. et al. Recurrent de novo mutations in PACS1 cause defective cranial-neural-crest migration and define a recognizable intellectual-disability syndrome. Am. J. Hum. Genet. 91, 1122–1127 (2012).
van der Maaten, L. & Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 9, 2579–2605 (2008).
Rousseeuw, P. J. Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. J. Comput. Appl. Math. 20, 53–65 (1987).
Ebstein, F. et al. De novo variants in the PSMC3 proteasome AAA-ATPase subunit gene cause neurodevelopmental disorders associated with type I interferonopathies. Preprint at medRxiv https://doi.org/10.1101/2021.12.07.21266342 (2021).
Richards, S. et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 17, 405–424 (2015).
Tavtigian, S. V. et al. Modeling the ACMG/AMP variant classification guidelines as a Bayesian classification framework. Genet. Med. 20, 1054–1060 (2018).
Philippakis, A. A. et al. The Matchmaker Exchange: a platform for rare disease gene discovery. Hum. Mutat. 36, 915–921 (2015).
Nguengang Wakap, S. et al. Estimating cumulative point prevalence of rare diseases: analysis of the Orphanet database. Eur. J. Hum. Genet. 28, 165–173 (2020).
Acknowledgements
This work was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) through individual grants to P.M.K. (grant nos. KR 3985/7-3, KR 3985/6-1). M.M.N. and R.C.B. are supported by the DFG through grants under the auspices of the Germany Excellence Strategy (grant nos. EXC2151–390873048, ImmunoSensation2). A. Schmidt received additional support by the BONFOR program of the Medical Faculty of the University of Bonn (grant no. 2020-1A-15). We also acknowledge support from the TRANSLATE-NAMSE project. We are also grateful for the language editing provided by N. Ruff.
Author information
Authors and Affiliations
Contributions
N.E., J.T.P., M.D., M.A.M., D.H., S.R., A.K., B.J., H.L., F.E., E.K., S.K., S.B., A. Schmidt, S.P., H.E., E.M., M.K., K.C., C.P., R.C.B., T. Bender, K.G.-H., T.B.H., M.W., T. Brunet, L.A., K.C.C., K.W.G. and G.J.L. collected and managed samples and data. T.-C.H., A.B.-H., G.B., A.H., H.K., S.S. and A. Schmid conducted data analysis. A.B.-H., G.B., T.K. and W.M. developed the software. N.E., K.W.G., D.H., N.F., H.B.B., M.S., C.P.S., S. Mundlos, S. Moosa, M.M.N. and P.M.K. provided intellectual input on clinical dysmorphology and translational, ethical and legal aspects. T.-C.H., A.B.-H., N.F., S. Moosa and P.M.K. wrote the manuscript with input from all authors. P.M.K. conceived and directed the study with input from all authors.
Corresponding author
Ethics declarations
Competing interests
A.B.-H., N.F. and G.B. are employees of FDNA. T.K. is an employee of GeneTalk GmbH. M.A.M. is a participant in the BIH Charité Digital Clinician Scientist Program founded by the late Prof. Duska Dragun and funded by the Charité-Universitätsmedizin Berlin and the Berlin Institute of Health. M.M.N. reported receiving personal fees from the Lundbeck Foundation, Robert Bosch Stiftung, Shire GmbH, Life & Brain GmbH and HMG Systems Engineering GmbH outside the submitted work. The other authors declare no conflicts of interest.
Peer review
Peer review information
Nature Genetics thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Performance improvement of double syndromes and double subjects when using different base sample sizes with Face2Gene models and the Face2Gene rare set.
Base sample size is calculated as the number of subjects multiplied by the number of syndromes. For example, the point of 40 subjects and 10 syndromes has sample size of 400, and it equals both the point of 10 subjects and 40 syndromes and the point of 20 subjects and 20 syndromes. ΔTop-10 accuracy is the difference of accuracy between the double syndromes or subjects and the base point, and is calculated based on Fig. 3. Take the two points annotated in the figure as two examples. The base point is 10 subjects and 40 syndromes with sample size 400. The upper indicated point is subtracting the point of 10 subjects and 40 syndromes from the point of 10 subjects and 80 syndromes in Fig. 3. The lower point is subtracting the point of 10 subjects and 40 syndromes from the point of 20 subjects and 40 syndromes in Fig. 3. In this graph, doubling the number of syndromes always improves top-10 accuracy more than doubling the number of subjects, particularly at larger base sample sizes. Thus, adding more syndromes is more effective than adding more subjects when enlarging the training set.
Extended Data Fig. 2 Influence of the number of syndromes included in model training.
The x-axis is the number of syndromes used in model training. The left y-axis shows the average top-10 accuracy for five models, and the error bars show the standard deviation over five models. The right y-axis is the cumulative number of subjects in the training syndromes. Each point is the average of testing five different models with different data splits. The null accuracy is 1.23% (10/816).
Extended Data Fig. 3 Comparison of the pairwise distance distribution between subjects in the same family and subjects in different families with the same disease-causing gene.
The median distance between affected individuals from the same family is 0.522, and the median distance between individuals from different families is 0.823. In the box plots, the center line indicates the median values, and the bottom and top edge of the box are the first (25%) and the third (75%) quartiles. The whiskers extend the data points outside the 1st to the 3rd quartiles. The total number of data points (n) for the same family is 28, and n is 928 for the different families.
Extended Data Fig. 4 Hierarchical clustering of four phenotypic series using a t-SNE projection of the Facial Phenotype Descriptors.
The projection shows clustering of FPDs for Kabuki syndrome, Noonan syndrome, mucopolysaccharidosis, and Cornelia de Lange syndrome.
Extended Data Fig. 5 t-SNE visualization of Facial Phenotype Descriptors of syndromes with or without facial dysmorphism.
a, Ten syndromes with facial dysmorphism. b, Ten syndromes without facial dysmorphism.
Extended Data Fig. 6 Screenshot of the GestaltMatcher web service.
Users can upload a patient photo to match against patients in the selected categories and can also visualize the clustering of patients by t-SNE. Access can be requested from www.gestaltmatcher.org. If the category DeepGestalt is selected, only cases with one of the frequent 299 diagnoses that DeepGestalt supports populate the gallery. If category Ultra-rare is chosen, the gallery is populated by cases with one of the 816 diagnoses not supported by DeepGestalt. The category of Undiagnosed Patients is suitable for a research setting if no match with a known disorder could be made (see, for example, PSMC3 in the online demo).
Extended Data Fig. 7 Overview of Face2Gene data categorization in GestaltMatcher.
The data were first divided by the number of subjects in each syndrome. Syndromes with more than six subjects were denoted frequent syndromes, and those with six or fewer as rare syndromes. Frequent syndromes were also recognized by DeepGestalt. Each category was further divided into a gallery and a test set. For each frequent syndrome, 90% of subjects were assigned to the gallery and used for model training; the remaining 10% of subjects were kept for validating the model training and were sampled in the test set. We performed 10-fold cross-validation on rare syndromes. In each syndrome, 90% of subjects were assigned to the gallery and 10% of subjects were assigned to the test set.
Extended Data Fig. 8 Venn diagram of numbers of syndromes in the Face2Gene and GMDB datasets.
Within each dataset, frequent syndromes are defined as those with seven or more subjects, and rare syndromes are defined as those with six or fewer subjects.
Supplementary information
Supplementary Information
Supplementary Note, Figs. 1–9 and Tables 1–7 and 9.
Supplementary Table
Supplementary Table 8. Summary of the Face2Gene and GMDB datasets.
Rights and permissions
About this article
Cite this article
Hsieh, TC., Bar-Haim, A., Moosa, S. et al. GestaltMatcher facilitates rare disease matching using facial phenotype descriptors. Nat Genet 54, 349–357 (2022). https://doi.org/10.1038/s41588-021-01010-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-021-01010-x
This article is cited by
-
Facial appearance associates with longitudinal multi-organ failure: an ICU cohort study
Critical Care (2024)
-
Potential of Artificial Intelligence to Accelerate Drug Development for Rare Diseases
Pharmaceutical Medicine (2024)
-
Kurze Wege zur Diagnose mit künstlicher Intelligenz – systematische Literaturrecherche zu „diagnostic decision support systems“
Der Schmerz (2024)
-
Coordinate-wise monotonic transformations enable privacy-preserving age estimation with 3D face point cloud
Science China Life Sciences (2024)
-
A diagnostic support system based on pain drawings: binary and k-disease classification of EDS, GBS, FSHD, PROMM, and a control group with Pain2D
Orphanet Journal of Rare Diseases (2023)