Abstract
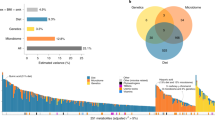
The gut microbiome has been implicated in a variety of physiological states, but controversy over causality remains unresolved. Here, we performed bidirectional Mendelian randomization analyses on 3,432 Chinese individuals with whole-genome, whole-metagenome, anthropometric and blood metabolic trait data. We identified 58 causal relationships between the gut microbiome and blood metabolites, and replicated 43 of them. Increased relative abundances of fecal Oscillibacter and Alistipes were causally linked to decreased triglyceride concentration. Conversely, blood metabolites such as glutamic acid appeared to decrease fecal Oxalobacter, and members of Proteobacteria were influenced by metabolites such as 5-methyltetrahydrofolic acid, alanine, glutamate and selenium. Two-sample Mendelian randomization with data from Biobank Japan partly corroborated results with triglyceride and with uric acid, and also provided causal support for published fecal bacterial markers for cancer and cardiovascular diseases. This study illustrates the value of human genetic information to help prioritize gut microbial features for mechanistic and clinical studies.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The human reference hg38 datasets are publicly available from http://hgdownload.soe.ucsc.edu/goldenPath/hg38/bigZips/. All summary statistics that support the findings of this study, including the associations between host genetics and microbiomes, host genetics and metabolites are publicly available from https://ftp.cngb.org/pub/CNSA/data2/CNP0000794/. The release of the data was approved by the Ministry of Science and Technology of China (Project ID: 2020BAT1137). Individual-level data including host genetics, metagenomics and metabolites have been uploaded to the GSA database (https://ngdc.cncb.ac.cn/bioproject/browse/PRJCA005334). Access to individual-level data has to be approved by corresponding authors (tao.zhang@genomics.cn, jiahuijue@genomics.cn), and is subject to the policies and approvals from the Human Genetic Resource Administration, Ministry of Science and Technology of the People’s Republic of China. The summary statistics data for 42 diseases and 59 blood quantitative traits in 212,453 Japanese individuals are available from Biobank Japan (http://jenger.riken.jp/en/result).
Code availability
The host genome reads were aligned to the latest reference human genome GRCh38/hg38 with BWA (v.0.7.15; http://bio-bwa.sourceforge.net/). The alignments were indexed in the BAM format using Samtools (v.0.1.18; http://samtools.sourceforge.net/) and PCR duplicates were marked for downstream filtering using Picardtools (v.1.62; http://broadinstitute.github.io/picard/). The genome variants calling were performed using GATK (v.3.8; https://gatk.broadinstitute.org/hc/en-us). The low-depth data were imputed using BEAGLE (v.5.0; https://faculty.washington.edu/browning/beagle/b5_0.html). The metagenome reads were aligned to hg38 using SOAP (v.2.22; http://soap.genomics.org.cn). Quality control, association analyses and GRS analyses were performed in PLINK (v.1.90; http://zzz.bwh.harvard.edu/plink/). We performed variance explained analysis using the REML model in GCTA (v.1.26.0; https://cnsgenomics.com/software/gcta) and one-sample MR analyses using the TSLS method in the AER package (v.1.2-9; https://www.rdocumentation.org/packages/AER/versions/1.2-9/topics/ivreg). Two-sample MR analyses were performed in GSMR (v.1.0.7; http://cnsgenomics.com/software/gsmr/) and TwoSampleMR (v.0.5.6; https://mrcieu.github.io/TwoSampleMR/). All statistics analyses and visualizations were performed in R (v.3.2.5; https://www.r-project.org).
References
Wang, J. & Jia, H. Metagenome-wide association studies: fine-mining the microbiome. Nat. Rev. Microbiol. 14, 508–522 (2016).
Moschen, A. R. et al. Lipocalin 2 protects from inflammation and tumorigenesis associated with gut microbiota alterations. Cell Host Microbe 19, 455–469 (2016).
Long, X. et al. Peptostreptococcus anaerobius promotes colorectal carcinogenesis and modulates tumour immunity. Nat. Microbiol. 4, 2319–2330 (2019).
Zhu, F. et al. Transplantation of microbiota from drug-free patients with schizophrenia causes schizophrenia-like abnormal behaviors and dysregulated kynurenine metabolism in mice. Mol. Psychiatry 25, 2905–2918 (2020).
Liu, X. et al. A genome-wide association study for gut metagenome in Chinese adults illuminates complex diseases. Cell Discov. 7, 9 (2021).
Bonder, M. J. et al. The effect of host genetics on the gut microbiome. Nat. Genet. 48, 1407–1412 (2016).
Wang, J. et al. Genome-wide association analysis identifies variation in vitamin D receptor and other host factors influencing the gut microbiota. Nat. Genet. 48, 1396–1406 (2016).
Turpin, W. et al. Association of host genome with intestinal microbial composition in a large healthy cohort. Nat. Genet. 48, 1413–1417 (2016).
Blekhman, R. et al. Host genetic variation impacts microbiome composition across human body sites. Genome Biol. 16, 191 (2015).
Rothschild, D. et al. Environment dominates over host genetics in shaping human gut microbiota. Nature 555, 210–215 (2018).
Burgess, S., Timpson, N. J., Ebrahim, S. & Davey Smith, G. Mendelian randomization: where are we now and where are we going? Int. J. Epidemiol. 44, 379–388 (2015).
Yang, Q., Lin, S. L., Kwok, M. K., Leung, G. M. & Schooling, C. M. The roles of 27 genera of human gut microbiota in ischemic heart disease, type 2 diabetes mellitus, and their risk factors: a Mendelian randomization study. Am. J. Epidemiol. 187, 1916–1922 (2018).
Sanna, S. et al. Causal relationships among the gut microbiome, short-chain fatty acids and metabolic diseases. Nat. Genet. 51, 600–605 (2019).
Sakaue, S. et al. Trans-biobank analysis with 676,000 individuals elucidates the association of polygenic risk scores of complex traits with human lifespan. Nat. Med. 26, 542–548 (2020).
Shin, S. Y. et al. An atlas of genetic influences on human blood metabolites. Nat. Genet. 46, 543–550 (2014).
Cox, A. J. et al. Association of SNPs in the UGT1A gene cluster with total bilirubin and mortality in the Diabetes Heart Study. Atherosclerosis 229, 155–160 (2013).
MacArthur, J. et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res. 45, D896–D901 (2017).
Verbanck, M., Chen, C. Y., Neale, B. & Do, R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 50, 693–698 (2018).
Katano, Y. et al. Complete genome sequence of Oscillibacter valericigenes Sjm18-20T (=NBRC 101213T). Stand. Genom. Sci. 6, 406–414 (2012).
Thingholm, L. B. et al. Obese individuals with and without type 2 diabetes show different gut microbial functional capacity and composition. Cell Host Microbe 26, 252–264.e10 (2019).
Hu, H. J. et al. Obesity alters the microbial community profile in Korean adolescents. PLoS One 10, e0134333 (2015).
Tims, S. et al. Microbiota conservation and BMI signatures in adult monozygotic twins. ISME J. 7, 707–717 (2013).
Noack, J., Dongowski, G., Hartmann, L. & Blaut, M. The human gut bacteria Bacteroides thetaiotaomicron and Fusobacterium varium produce putrescine and spermidine in cecum of pectin-fed gnotobiotic rats. J. Nutr. 130, 1225–1231 (2000).
Luis, A. S. et al. Dietary pectic glycans are degraded by coordinated enzyme pathways in human colonic Bacteroides. Nat. Microbiol. 3, 210–219 (2018).
PeBenito, A. et al. Comparative prevalence of Oxalobacter formigenes in three human populations. Sci. Rep. 9, 574 (2019).
Ishigaki, K. et al. Large-scale genome-wide association study in a Japanese population identifies novel susceptibility loci across different diseases. Nat. Genet. 52, 669–679 (2020).
Huo, Y. et al. Efficacy of folic acid therapy in primary prevention of stroke among adults with hypertension in China: the CSPPT randomized clinical trial. JAMA 313, 1325–1335 (2015).
Jie, Z. et al. The gut microbiome in atherosclerotic cardiovascular disease. Nat. Commun. 8, 845 (2017).
Yachida, S. et al. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat. Med. 25, 968–976 (2019).
Reese, A. T. et al. Microbial nitrogen limitation in the mammalian large intestine. Nat. Microbiol. 3, 1441–1450 (2018).
Petrus, P. et al. Glutamine links obesity to inflammation in human white adipose tissue. Cell Metab. 31, 375–390.e11 (2019).
Liu, R. et al. Gut microbiome and serum metabolome alterations in obesity and after weight-loss intervention. Nat. Med. 23, 859–868 (2017).
Choi, W. M. et al. Glutamate signaling in hepatic stellate cells drives alcoholic steatosis. Cell Metab. 30, 877–889.e877 (2019).
Kang, D. J. et al. Gut microbiota drive the development of neuroinflammatory response in cirrhosis in mice. Hepatology 64, 1232–1248 (2016).
Long, T. et al. Plasma metals and cardiovascular disease in patients with type 2 diabetes. Environ. Int. 129, 497–506 (2019).
Kuo, C. F., Grainge, M. J., Zhang, W. & Doherty, M. Global epidemiology of gout: prevalence, incidence and risk factors. Nat. Rev. Rheumatol. 11, 649–662 (2015).
Li, J. et al. An integrated catalog of reference genes in the human gut microbiome. Nat. Biotechnol. 32, 834–841 (2014).
Qin, J. et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65 (2010).
Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012).
Maier, L. et al. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature 555, 623–628 (2018).
Polderman, T. J. et al. Meta-analysis of the heritability of human traits based on fifty years of twin studies. Nat. Genet. 47, 702–709 (2015).
Yang, J., Lee, S. H., Goddard, M. E. & Visscher, P. M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88, 76–82 (2011).
Jie, Z. et al. A transomic cohort as a reference point for promoting a healthy human gut microbiome. Med. Microecol. 8, 100039 (2021).
Zhu, J. et al. Over 50,000 metagenomically assembled draft genomes for the human oral microbiome reveal new taxa. Genomics Proteom. Bioinform. https://doi.org/10.1016/j.gpb.2021.05.001 (2021).
Liu, X. et al. Metagenome-genome-wide association studies reveal human genetic impact on the oral microbiome. Preprint at bioRxiv https://doi.org/10.1101/2021.05.06.443017 (2021).
Fang, C. et al. Assessment of the cPAS-based BGISEQ-500 platform for metagenomic sequencing. Gigascience 7, 1–8 (2018).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Browning, B. L., Zhou, Y. & Browning, S. R. A one-penny imputed genome from next-generation reference panels. Am. J. Hum. Genet. 103, 338–348 (2018).
Li, R. et al. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25, 1966–1967 (2009).
Vieira-Silva, S. et al. Species-function relationships shape ecological properties of the human gut microbiome. Nat. Microbiol. 1, 16088 (2016).
Caspi, R. et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases. Nucleic Acids Res. 42, D459–D471 (2014).
Teumer, A. Common methods for performing Mendelian randomization. Front. Cardiovasc. Med. 5, 51 (2018).
Permutt, T. & Hebel, J. R. Simultaneous-equation estimation in a clinical trial of the effect of smoking on birth weight. Biometrics 45, 619–622 (1989).
Zhu, Z. et al. Causal associations between risk factors and common diseases inferred from GWAS summary data. Nat. Commun. 9, 224 (2018).
Burgess, S., Butterworth, A. & Thompson, S. G. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet. Epidemiol. 37, 658–665 (2013).
Bowden, J. et al. A framework for the investigation of pleiotropy in two-sample summary data Mendelian randomization. Stat. Med. 36, 1783–1802 (2017).
Bowden, J., Davey Smith, G. & Burgess, S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 44, 512–525 (2015).
Bowden, J. et al. Assessing the suitability of summary data for two-sample Mendelian randomization analyses using MR-Egger regression: the role of the I2 statistic. Int. J. Epidemiol. 45, 1961–1974 (2016).
Bowden, J., Davey Smith, G., Haycock, P. C. & Burgess, S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet. Epidemiol. 40, 304–314 (2016).
Hartwig, F. P., Davey Smith, G. & Bowden, J. Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int. J. Epidemiol. 46, 1985–1998 (2017).
Acknowledgements
We are sincerely grateful for the support provided by China National GeneBank. We thank all the volunteers for their time and for self-collecting the fecal samples using our kit. We are very grateful to K. Kristiansen (Department of Biology, University of Copenhagen, Denmark; BGI-Qingdao, BGI-Shenzhen, China) for his support for the joint PhD program.
Author information
Authors and Affiliations
Contributions
H.J. and T.Z. conceived and organized this study. J.W. initiated the overall health project. X.X., H.Y., S.Z., Y.H., W.L. and Y. Zong contributed to organization of the cohort sample collection and questionnaire collection. H.L. led the DNA extraction and sequencing. X.Q., J.Z. and R.W. generated the metabolic data. X. Liu, T.Z. and X.T. processed the whole-genome data. Y. Zou, X. Lin, Z.Z., H.Z., L.T., Q.W., Z.J. and L.X. processed the metagenome data. X. Liu and X.T. performed the Mendelian randomization analyses. X. Liu and H.J. wrote the manuscript. All authors contributed to the data and text in this manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Genetics thanks Yukinori Okada and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Notes and Figs. 1–8.
Supplementary Tables
Supplementary Tables 1–16.
Rights and permissions
About this article
Cite this article
Liu, X., Tong, X., Zou, Y. et al. Mendelian randomization analyses support causal relationships between blood metabolites and the gut microbiome. Nat Genet 54, 52–61 (2022). https://doi.org/10.1038/s41588-021-00968-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-021-00968-y
This article is cited by
-
Inflammatory cytokines and their potential role in Sjogren’s syndrome risk: insights from a mendelian randomization study
Advances in Rheumatology (2024)
-
Blood lipids mediate the effects of gut microbiome on endometriosis: a mendelian randomization study
Lipids in Health and Disease (2024)
-
Causal relationship between gut microbiota and gastrointestinal diseases: a mendelian randomization study
Journal of Translational Medicine (2024)
-
Univariable and multivariable Mendelian randomization study identified the key role of gut microbiota in immunotherapeutic toxicity
European Journal of Medical Research (2024)
-
Putative causal relations among gut flora, serums metabolites and arrhythmia: a Mendelian randomization study
BMC Cardiovascular Disorders (2024)