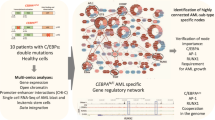
Abstract
Altered transcription is a cardinal feature of acute myeloid leukemia (AML); however, exactly how mutations synergize to remodel the epigenetic landscape and rewire three-dimensional DNA topology is unknown. Here, we apply an integrated genomic approach to a murine allelic series that models the two most common mutations in AML: Flt3-ITD and Npm1c. We then deconvolute the contribution of each mutation to alterations of the epigenetic landscape and genome organization, and infer how mutations synergize in the induction of AML. Our studies demonstrate that Flt3-ITD signals to chromatin to alter the epigenetic environment and synergizes with mutations in Npm1c to alter gene expression and drive leukemia induction. These analyses also allow the identification of long-range cis-regulatory circuits, including a previously unknown superenhancer of Hoxa locus, as well as larger and more detailed gene-regulatory networks, driven by transcription factors including PU.1 and IRF8, whose importance we demonstrate through perturbation of network members.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
All sequencing raw data, normalized bigwig tracks for RNA-seq, ChIP-seq and ATAC-seq have been deposited in the GEO database under the series GSE146669 (subseries GSE146668 for RNA-seq, GSE146663 for ChIP-seq, GSE146613 for ATAC-seq and GSE146662 for pCHiC) and with no restrictions to access. All supporting data derived from the sequencing analysis to assist understanding of the results and discussions in the paper were provided in several supplementary tables. The studies have also reanalyzed multiple datasets that are publicly available: the ChIP-seq on BRD4 (ArrayExpress: ERR220396) and H3K27ac (GSM2716711) in OCI-AML3 cells; H3K27ac in Kasumi-1 cells (GSM2212053); H3K4me1 (GSM1816068) and CTCF (GSM651541) in human CD34+ HSPC; the DHS-seq in AML patients (GSM2893610, GSM2893614, GSM2893615 and GSM2893616); the ATAC-seq in human CD34+ HSPC (GSM1888536); the pCHiC from human CD34+ cells (ArrayExpress: ERR436027); H3K4me1 (GSM14412890), H3K27ac (GSM1441273) and ATAC-seq (GSM1463173) in mouse GMP cells. Source data are provided with this paper.
Code availability
All computational analysis is described in Methods with either the software default parameters and pipelines or custom code available at https://github.com/haiyang-yun/3D_chromatin_in_AML (archived also on Zenodo: https://doi.org/10.5281/zenodo.5009065)78.
References
Maston, G. A., Evans, S. K. & Green, M. R. Transcriptional regulatory elements in the human genome. Annu Rev. Genomics Hum. Genet 7, 29–59 (2006).
Lagha, M., Bothma, J. P. & Levine, M. Mechanisms of transcriptional precision in animal development. Trends Genet. 28, 409–416 (2012).
Spitz, F. & Furlong, E. E. Transcription factors: from enhancer binding to developmental control. Nat. Rev. Genet. 13, 613–626 (2012).
Schoenfelder, S. & Fraser, P. Long-range enhancer-promoter contacts in gene expression control. Nat. Rev. Genet. 20, 437–455 (2019).
Laurenti, E. & Gottgens, B. From haematopoietic stem cells to complex differentiation landscapes. Nature 553, 418–426 (2018).
Huang, J. et al. Dynamic control of enhancer repertoires drives lineage and stage-specific transcription during hematopoiesis. Dev. Cell 36, 9–23 (2016).
Orkin, S. H. & Zon, L. I. Hematopoiesis: an evolving paradigm for stem cell biology. Cell 132, 631–644 (2008).
Cedar, H. & Bergman, Y. Epigenetics of haematopoietic cell development. Nat. Rev. Immunol. 11, 478–488 (2011).
Cancer Genome Atlas Research, N. et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 368, 2059–2074 (2013).
Grimwade, D., Ivey, A. & Huntly, B. J. Molecular landscape of acute myeloid leukemia in younger adults and its clinical relevance. Blood 127, 29–41 (2016).
Papaemmanuil, E. et al. Genomic classification and prognosis in acute myeloid leukemia. N. Engl. J. Med. 374, 2209–2221 (2016).
Dohner, H., Weisdorf, D. J. & Bloomfield, C. D. Acute myeloid leukemia. N. Engl. J. Med. 373, 1136–1152 (2015).
Tenen, D. G. Disruption of differentiation in human cancer: AML shows the way. Nat. Rev. Cancer 3, 89–101 (2003).
Lee, B. H. et al. FLT3 mutations confer enhanced proliferation and survival properties to multipotent progenitors in a murine model of chronic myelomonocytic leukemia. Cancer Cell 12, 367–380 (2007).
Vassiliou, G. S. et al. Mutant nucleophosmin and cooperating pathways drive leukemia initiation and progression in mice. Nat. Genet. 43, 470–475 (2011).
Mupo, A. et al. A powerful molecular synergy between mutant Nucleophosmin and Flt3-ITD drives acute myeloid leukemia in mice. Leukemia 27, 1917–1920 (2013).
Buenrostro, J. D. et al. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013).
Mifsud, B. et al. Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat. Genet. 47, 598–606 (2015).
Assi, S. A. et al. Subtype-specific regulatory network rewiring in acute myeloid leukemia. Nat. Genet. 51, 151–162 (2019).
Bell, O., Tiwari, V. K., Thoma, N. H. & Schubeler, D. Determinants and dynamics of genome accessibility. Nat. Rev. Genet. 12, 554–564 (2011).
Snow, J. W. et al. Context-dependent function of ‘GATA switch’ sites in vivo. Blood 117, 4769–4772 (2011).
Yamazaki, H. et al. A remote GATA2 hematopoietic enhancer drives leukemogenesis in inv(3)(q21;q26) by activating EVI1 expression. Cancer Cell 25, 415–427 (2014).
Baek, S., Goldstein, I. & Hager, G. L. Bivariate genomic footprinting detects changes in transcription factor activity. Cell Rep. 19, 1710–1722 (2017).
Calo, E. & Wysocka, J. Modification of enhancer chromatin: what, how, and why? Mol. Cell 49, 825–837 (2013).
Lara-Astiaso, D. et al. Immunogenetics. Chromatin state dynamics during blood formation. Science 345, 943–949 (2014).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Lieberman-Aiden, E. et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 (2009).
Tischkowitz, M. et al. Bi-allelic silencing of the Fanconi anaemia gene FANCF in acute myeloid leukaemia. Br. J. Haematol. 123, 469–471 (2003).
Freire-Pritchett, P. et al. Global reorganisation of cis-regulatory units upon lineage commitment of human embryonic stem cells. eLife 6, 21926 (2017).
Rubin, A. J. et al. Lineage-specific dynamic and pre-established enhancer-promoter contacts cooperate in terminal differentiation. Nat. Genet. 49, 1522–1528 (2017).
Ma, Q. et al. Super-Enhancer redistribution as a mechanism of broad gene dysregulation in repeatedly drug-treated cancer cells. Cell Rep. 31, 107532 (2020).
Satija, R. et al. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495–502 (2015).
Li, Y. et al. Regulation of the PU.1 gene by distal elements. Blood 98, 2958–2965 (2001).
Okuno, Y. et al. Potential autoregulation of transcription factor PU.1 by an upstream regulatory element. Mol. Cell. Biol. 25, 2832–2845 (2005).
Rosenbauer, F. et al. Lymphoid cell growth and transformation are suppressed by a key regulatory element of the gene encoding PU.1. Nat. Genet. 38, 27–37 (2006).
Will, B. et al. Minimal PU.1 reduction induces a preleukemic state and promotes development of acute myeloid leukemia. Nat. Med. 21, 1172–1181 (2015).
Hu, G. et al. Regulation of nucleosome landscape and transcription factor targeting at tissue-specific enhancers by BRG1. Genome Res. 21, 1650–1658 (2011).
Mazumdar, C. et al. Leukemia-associated cohesin mutants dominantly enforce stem cell programs and impair human hematopoietic progenitor differentiation. Cell Stem Cell 17, 675–688 (2015).
Dawson, M. A. et al. Recurrent mutations, including NPM1c, activate a BRD4-dependent core transcriptional program in acute myeloid leukemia. Leukemia 28, 311–320 (2014).
Zhao, Y. et al. High-resolution mapping of RNA polymerases identifies mechanisms of sensitivity and resistance to BET inhibitors in t(8;21) AML. Cell Rep. 16, 2003–2016 (2016).
Gerlach, D. et al. The novel BET bromodomain inhibitor BI 894999 represses super-enhancer-associated transcription and synergizes with CDK9 inhibition in AML. Oncogene 37, 2687–2701 (2018).
Dovey, O. M. et al. Molecular synergy underlies the co-occurrence patterns and phenotype of NPM1-mutant acute myeloid leukemia. Blood 130, 1911–1922 (2017).
Foletta, V. C., Segal, D. H. & Cohen, D. R. Transcriptional regulation in the immune system: all roads lead to AP-1. J. Leukoc. Biol. 63, 139–152 (1998).
Madrigal, P. & Alasoo, K. AP-1 takes centre stage in enhancer chromatin dynamics. Trends Cell Biol. 28, 509–511 (2018).
Carotta, S., Wu, L. & Nutt, S. L. Surprising new roles for PU.1 in the adaptive immune response. Immunol. Rev. 238, 63–75 (2010).
Voon, D. C., Hor, Y. T. & Ito, Y. The RUNX complex: reaching beyond haematopoiesis into immunity. Immunology 146, 523–536 (2015).
Cauchy, P. et al. Chronic FLT3-ITD Signaling in acute myeloid leukemia is connected to a specific chromatin signature. Cell Rep. 12, 821–836 (2015).
Gadad, S. S. et al. The multifunctional protein nucleophosmin (NPM1) is a human linker histone H1 chaperone. Biochemistry 50, 2780–2789 (2011).
Nemeth, A. et al. Initial genomics of the human nucleolus. PLoS Genet. 6, e1000889 (2010).
van Koningsbruggen, S. et al. High-resolution whole-genome sequencing reveals that specific chromatin domains from most human chromosomes associate with nucleoli. Mol. Biol. Cell 21, 3735–3748 (2010).
Yan, J. et al. Histone H3 lysine 4 monomethylation modulates long-range chromatin interactions at enhancers. Cell Res. 28, 204–220 (2018).
Tzelepis, K. et al. A CRISPR dropout screen identifies genetic vulnerabilities and therapeutic targets in acute myeloid leukemia. Cell Rep. 17, 1193–1205 (2016).
Wang, T. et al. Gene essentiality profiling reveals gene networks and synthetic lethal interactions with oncogenic ras. Cell 168, 890–903 e815 (2017).
McKenzie, M. D. et al. Interconversion between tumorigenic and differentiated states in acute myeloid leukemia. Cell Stem Cell 25, 258–272 e259 (2019).
Rosenbauer, F., Koschmieder, S., Steidl, U. & Tenen, D. G. Effect of transcription−factor concentrations on leukemic stem cells. Blood 106, 1519–1524 (2005).
Bonadies, N., Pabst, T. & Mueller, B. U. Heterozygous deletion of the PU.1 locus in human AML. Blood 115, 331–334 (2010).
Huang, G. et al. The ability of MLL to bind RUNX1 and methylate H3K4 at PU.1 regulatory regions is impaired by MDS/AML-associated RUNX1/AML1 mutations. Blood 118, 6544–6552 (2011).
Vangala, R. K. et al. The myeloid master regulator transcription factor PU.1 is inactivated by AML1-ETO in t(8;21) myeloid leukemia. Blood 101, 270–277 (2003).
Bell, C. C. et al. Targeting enhancer switching overcomes non-genetic drug resistance in acute myeloid leukaemia. Nat. Commun. 10, 2723 (2019).
Cusan, M. et al. LSD1 inhibition exerts its antileukemic effect by recommissioning PU.1− and C/EBPalpha-dependent enhancers in AML. Blood 131, 1730–1742 (2018).
Gozdecka, M. et al. UTX-mediated enhancer and chromatin remodeling suppresses myeloid leukemogenesis through noncatalytic inverse regulation of ETS and GATA programs. Nat. Genet. 50, 883–894 (2018).
Behre, G. et al. Meropenem monotherapy versus combination therapy with ceftazidime and amikacin for empirical treatment of febrile neutropenic patients. Ann. Hematol. 76, 73–80 (1998).
Grondin, B. et al. c-Jun homodimers can function as a context-specific coactivator. Mol. Cell. Biol. 27, 2919–2933 (2007).
Huang, Y. et al. Identification and characterization of Hoxa9 binding sites in hematopoietic cells. Blood 119, 388–398 (2012).
Zhou, J. et al. PU.1 is essential for MLL leukemia partially via crosstalk with the MEIS/HOX pathway. Leukemia 28, 1436–1448 (2014).
Neijts, R. & Deschamps, J. At the base of colinear Hox gene expression: cis-features and trans-factors orchestrating the initial phase of Hox cluster activation. Dev. Biol. 428, 293–299 (2017).
Noordermeer, D. et al. The dynamic architecture of Hox gene clusters. Science 334, 222–225 (2011).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Conesa, A. et al. A survey of best practices for RNA-seq data analysis. Genome Biol. 17, 13 (2016).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Bailey, T. et al. Practical guidelines for the comprehensive analysis of ChIP–seq data. PLoS Comput. Biol. 9, e1003326 (2013).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Ross-Innes, C. S. et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature 481, 389–393 (2012).
Ramirez, F. et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016).
Wingett, S. et al. HiCUP: pipeline for mapping and processing Hi-C data. F1000Res 4, 1310 (2015).
Cairns, J. et al. CHiCAGO: robust detection of DNA looping interactions in Capture Hi-C data. Genome Biol. 17, 127 (2016).
Yun, H. haiyang-yun/3D_chromatin_in_AML. Zenodo https://doi.org/10.5281/zenodo.5009065 (2021).
Acknowledgements
This study was carried out in the laboratory of B.J.P.H. with funding from Cancer Research UK (C18680/A25508), the European Research Council (647685), MRC (MR-R009708-1), the Kay Kendall Leukaemia Fund (KKL1243), the Wellcome Trust (205254/Z/16/Z) and the Cancer Research UK Cambridge Major Centre (C49940/A25117). This research was supported by the NIHR Cambridge Biomedical Research Centre (BRC-1215-20014), and was funded in part by the Wellcome Trust, who supported the Wellcome – MRC Cambridge Stem Cell Institute (203151/Z/16/Z) and Cambridge Institute for Medical Research (100140/Z/12/Z). The views expressed are those of the authors and not necessarily those of the NIHR or the Department of Health and Social Care. D.L.-A. is Marie-Skłodowska-Curie International Fellow (886474). D.S. was a postdoctoral fellow of the Mildred-Scheel Organization, German Cancer Aid (111875). P.G. is funded by a Cancer Research UK Advanced Clinician Scientist Fellowship (C57799/A27964) and was previously funded by a Wellcome Trust fellowship (109967/Z/15/Z) and an ASH Global Research Award. G.S.V. is a CRUK Senior Cancer Research Fellow (C22/324/A23015) and work in his laboratory was supported by Blood Cancer UK (17006). For the purpose of Open Access, the author has applied a CC BY public copyright licence to any Author Accepted Manuscript version arising from this submission. We acknowledge M. Dawes in the Department of Hematology for interlab communications and organizational assistance, and thank M. Paramor in the NGS library facility at Cambridge Stem Cell Institute for the help with preparation of RNA-seq libraries and the Cancer Research UK (CRUK) Cambridge Institute Genomics Core for providing the NGS services.
Author information
Authors and Affiliations
Contributions
H.Y. and B.J.P.H. conceived the study, designed the experiments and prepared the manuscript. H.Y. designed and conducted most of the experiments and analyzed the data. N.N. performed the CRISPR experiments in human leukemic cell lines. S.V. executed the NGS data alignment and helped with bioinformatic analysis. G.G. performed the flow cytometry experiments. A.M. and G.S.V. coordinated mouse tissues. P.M. performed the BaGFoot analysis. D.S., D.L.-A., S.J.H., S.A.-S., F.B., L.M., A.C.-V. and P.G. provided technical assistance. E.M. and X.W. helped with computational analysis. M.G. and O.M.D. provided the DM-Cas9 cells. C.M.-T. participated in the discussion of the project. C.S.O. provided expertise on promoter capture HiC assays. B.J.P.H supervised all the work and directed the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Genetics thanks Charles Mullighan and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Transcriptional changes across WT and mutant HSPC.
a, Heatmap showing unsupervised clustering analysis of global gene expression in WT and mutant HSPC. b, Top 10 positively enriched gene ontology (GO) gene sets revealed by GSEA for each mutant (vs WT). c, Commonly enriched gene sets from GSEA analysis of differential gene expression by Npm1c and DM. d, GSEA enrichment plots showing gene set of Kong_E2F3_Targets in Npm1c or DM vs WT by GSEA analysis. e, Gene sets positively or negatively enriched in all three mutants vs WT.
Extended Data Fig. 2 Global chromatin accessibility across WT and mutant HSPC.
a, Heatmaps and profile plots of ATAC-seq enrichment across WT and mutant HSPC over regions with gain or loss of accessibility in the presence of Npm1c. Peaks were ranked by average enrichment across all samples. b, Chromatin accessibility at the Gata2 genes and its upstream enhancers in all four HSPC and wildtype neutrophils. Regions showing loss of accessibility in Npmc1 and DM HSPC are highlighted. c, Linking differential accessibility at gene promoters to their mRNA expression changes by each mutant. Up- or downregulation were defined by setting adjP (two-tailed and multiple testing corrected) < 0.05 and absolute FC ≥ 1.5. d-f, De novo motifs significantly enriched at genomic regions with altered accessibility by Npm1c (d) and Flt3-ITD (e) or open chromatin sites with static accessibility (f) where FDR of differential analysis > 0.2 in any mutant vs WT HSPC. HOMER outputs motifs with target coverage > 10% and ranked by p values (one-tailed, not multiple testing corrected). g and h, BaGFoot analysis illustrates TFs with differential footprint depth and accessibility in Npm1c (g) and Flt3-ITD (h) vs WT HSPC. Motifs outside the fence and with a p value (two-tailed, not multiple testing corrected) < 0.05 are statistically significant outliers; n.s., not significant.
Extended Data Fig. 3 Chromatin modifications at accessible enhancers across WT and mutant HSPC.
a, Distribution of all H3K4me3 peaks based on read counts across all four cell types. Active promoters were marked by high H3K4me3 with log2 CPM > 4. b, Genome distribution of ATAC-seq consensus peaks. c, Number of total accessible enhancer peaks defined in all four HSPC samples and percentage of dynamic enhancers which were defined by differential H3K4me1 in the presence of any mutations. d, Numbers of overlapping accessible enhancers with gain or loss of H3K4me1 in the presence of Flt3-ITD or DM. Hypergeometric test p values (one-tailed, not multiple testing corrected) are shown. e, Heatmaps of H3K4me1 enrichment over gained or lost enhancers during DM leukemia induction. Peaks were ranked by average enrichment across all samples. f, Percentage of accessible enhancers marked by both H3K4me1 and H3K27ac in total accessible enhancers across all four HSPC states. g, Percentage of enhancers showing dynamic H3K27ac modification in the presence of single or double mutations. h. Heatmaps and profile plots of H3K27ac enrichment over enhancers showing gain or loss of H3K27ac during DM leukemia induction. Peaks were ranked by average enrichment across all samples.
Extended Data Fig. 4 Immunophenotypic characterization of HSPCs isolated from bone marrow of wildtype and mutant mice.
a, Representative flow cytometry plots showing proportions of hematopoietic stem and progenitors within bone marrow compartment. Lin-, lineage negative; HPC, hematopoietic progenitors (Lin−Sca1−cKit+) containing all myeloid progenitors; LSK, Lin−Sca1+cKit+; CMP, common myeloid progenitors; GMP, granulocyte/macrophage progenitors; MEP, megakaryocyte/erythroid progenitors. b, Comparison of proportions of hematopoietic stem and progenitors in their parental populations in mutant mice (Npm1flox-cA/+;Mx1-Cre, n = 9; Flt3ITD/+, n = 8; Npm1flox-cA/+;Mx1-Cre;Flt3ITD/+, n = 9) compared with wildtype mice (WT, n = 7). Student’s unpaired t-tests (two-sided) were performed for the comparisons; data are presented as mean values ± standard deviation; Only the p values < 0.05 were shown.
Extended Data Fig. 5 3D chromatin interaction profiles across WT and mutant HSPC.
a, Numbers of total and high-confidence pCHiC interactions in all four HSPC states. High-confidence interactions were defined as significant interactions (with CHiCAGO score ≥ 5) overlapping in both replicates of each cell type. b, Numbers of pCHiC interactions captured by individual promoters. c, Distance of interacting regions from their target promoters. Median distance was shown. d, Illustration of chromatin compartments A/B levels at a DM-induced ‘B to A’ flipped region (highlighted) containing the Igf1 oncogene. e. Illustration of chromatin compartments A/B levels at a DM-induced ‘B to A’ flipped region (highlighted) containing an Ifi (interferon-inducible) gene cluster (in red). f and g, Significant interactions, represented by arcs, associated with the Gfi1b (f) or Irf8 (g) promoters in different cell types.
Extended Data Fig. 6 Integrative analysis with Seurat-guided clustering of chromatin profiles across WT and mutant HSPC.
a, Profile plots of chromatin marks and accessibility at accessible regions in each of the 10 clusters (except for Cluster-6 and -10). b, De novo motifs significantly enriched at Clusters 7–9 chromatin regions. HOMER outputs motifs with target coverage > 10% and ranked by p values (one-tailed, not multiple testing corrected). c, GO analysis of genes that are linked to the chromatin regions in Cluster-10 using pCHiC profiles and downregulated during leukemia induction. d, Volcano plot showing differential expression of Cluster-10 linked genes in DM vs WT HSPC. Up- or downregulation were defined by setting adjP (two-tailed and multiple testing corrected) < 0.05 and absolute FC ≥ 1.5. e, Percentage of Cluster-6 genes with human overlap showing their expression in DM vs WT HSPC. Differential expression was defined by setting adjP (two-tailed and multiple testing corrected) < 0.05 and absolute FC ≥ 1.5.
Extended Data Fig. 7 Correlating enhancer alterations with differential expression of target genes or DNA topology changes.
a, Percentage of dynamic enhancers interacting with gene promoters and the percent of up- or downregulation of the genes they linked to. Dynamic enhancers were marked by either differential H3K4me1 only, or together with differential H3K27ac and accessibility. b, Percentage of genes involving flipped chromatin compartments and expressed in DM vs WT HSPC. Differential expression was defined by setting adjP (two-tailed and multiple testing corrected) < 0.05 and absolute FC ≥ 1.5. c, Percentage of enhancers with dynamic interactions showing their H3K4me1 read counts in DM vs WT HSPC. Differential H3K4m1 levels were defined by setting FDR (two-tailed and multiple testing corrected) < 0.05 and FC ≥ 1.5. d, Example genomic region demonstrating correlation of H3K4me1 changes with ‘rewired’ interactions.
Extended Data Fig. 8 Differential Super-enhancer usage across WT and mutant HSPC.
a, Definition of super-enhancers (SEs) by ranking H3K27ac peaks that were overlapped in both replicates of each cell type based on normalised H3K27 counts. The top-ranked 801 regions were considered as SEs across four HSPC. b, Number of super-enhancers (SE) with increase (UP) or decrease (DOWN) in H3K27ac modification in mutant vs WT HSPC. c, Heatmaps and profile plots of H3K27ac enrichment in WT and mutant HSPC over SEs showing gain or loss of H3K27ac during DM leukemia induction. d, Linking differential activity of SEs to altered mRNA expression of their target genes (determined by pCHiC interactions) in mutant vs WT HSPC. Upregulated genes connecting to SEs with H3K27ac gain or downregulated genes connecting to SEs with H3K27ac loss were determined by setting adjP (two-tailed and multiple testing corrected) < 0.05 and FC ≥ 1.5 for expression, or FDR (two-tailed and multiple testing corrected) < 0.05 and FC ≥ 1.5 for H3K27ac. Several Hoxa genes were indicated in Npm1c vs WT. e, Significantly enriched GO terms for genes linked to gained SEs and upregulated in DM vs WT (upper panel) or for genes linked to lost SEs and downregulated (lower panel). f and g, Promoter-contact plots showing the read counts of promoter bait to target pairs (bait-‘other end’) for Spi1 (f) and Hoxa9/Hoxa10 (g). Regions highlighted yellow show rewired interactions.
Extended Data Fig. 9 Perturbation of critical cis-regulatory hubs and their target genes abrogates leukemia maintenance in mouse DM leukemia.
a, mRNA expression of AP-1 components (Jun, Fos) detected by RT-qPCR in DM cells expressing shRNAs targeting Jun or Fos relative to control shRNA (n = 3 independent experiments). b, CFU assays of DM cells expressing shRNAs targeting Jun or Fos or control gRNAs (n = 3 independent experiments). c, mRNA expression of Hoxa9, Hoxa10, and Spi1 in DM cells expressing shRNAs targeting them specifically relative to control shRNA (n = 3 independent experiments). d and e, CFU assays (d) and ex vivo cell proliferation (e) of DM cells expressing shRNAs targeting Hoxa9, Hoxa10, Spi1 or control shRNA (n = 3 independent experiments). f and i, Gel electrophoresis of PCR products on genomic DNA of DM-Cas9 cells expressing control gRNAs, Spi1-URE (f) or Hoxa-LRSE (i) gRNAs. PCR experiment was performed once on three independent gRNA transductions (marked with #). g and j, Sanger sequencing to confirm the deletion of Spi1-URE (g) or Hoxa-LRSE (j) in DM-Cas9 cells expressing Spi1-URE gRNAs. h and k, Ex vivo cell growth of DM-Cas9 cells expressing Spi1-URE gRNAs (h) or Hoxa-LRSE gRNAs (k) in comparison to cells expressing control gRNAs (n = 3 independent experiments). Statistical analyses in a-d, h and k were performed by running Student’s unpaired t-tests and p values (two-sided, not multiple testing corrected) were shown; error bars represent mean and standard error of mean.
Extended Data Fig. 10 Perturbation of critical target genes impairs human leukemia cells.
a and b, Relative mRNA expression of target gene expression upon CRISPR-mediated knockdown in OCI-AML3 (a) and MOLM-13 (b) leukemia cell lines (n = 3 independent experiments). c-e, Cell proliferation of MOLM-13 cells expressing human gRNAs targeting SPI1 (c), IRF8 (d) and IGF1 (e) relative to control gRNAs (n = 3, 4, 3 independent experiments, respectively). # indicates two independent gRNAs. f, CFU assays of MOLM-13 cells expressing human gRNAs targeting SPI1, IRF8 and IGF1 relative to control gRNAs (n = 3 independent experiments). Statistical analyses in a-f were performed by running Student’s unpaired t-tests and p values (two-sided, not multiple testing corrected) were shown; error bars represent mean and standard error of mean.
Supplementary information
Supplementary Information
Supplementary Note and Figs. 1–3.
Supplementary Tables
Supplementary Tables 1–13
Source data
Source Data
Raw gel electrophoresis image for Extended Data Fig. 9f,i.
Rights and permissions
About this article
Cite this article
Yun, H., Narayan, N., Vohra, S. et al. Mutational synergy during leukemia induction remodels chromatin accessibility, histone modifications and three-dimensional DNA topology to alter gene expression. Nat Genet 53, 1443–1455 (2021). https://doi.org/10.1038/s41588-021-00925-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-021-00925-9
This article is cited by
-
A new genomic framework to categorize pediatric acute myeloid leukemia
Nature Genetics (2024)
-
A novel network pharmacology approach for leukaemia differentiation therapy using Mogrify®
Oncogene (2022)