Abstract
Driver mutations in genes encoding histone H3 proteins resulting in p.Lys27Met substitutions (H3-K27M) are frequent in pediatric midline brain tumors. However, the precise mechanisms by which H3-K27M causes tumor initiation remain unclear. Here, we use human hindbrain neural stem cells to model the consequences of H3.3-K27M on the epigenomic landscape in a relevant developmental context. Genome-wide mapping of epitope-tagged histone H3.3 revealed that both the wild type and the K27M mutant incorporate abundantly at pre-existing active enhancers and promoters, and to a lesser extent at Polycomb repressive complex 2 (PRC2)-bound regions. At active enhancers, H3.3-K27M leads to focal H3K27ac loss, decreased chromatin accessibility and reduced transcriptional expression of nearby neurodevelopmental genes. In addition, H3.3-K27M deposition at a subset of PRC2 target genes leads to increased PRC2 and PRC1 binding and augmented transcriptional repression that can be partially reversed by PRC2 inhibitors. Our work suggests that, rather than imposing de novo transcriptional circuits, H3.3-K27M drives tumorigenesis by locking initiating cells in their pre-existing, immature epigenomic state, via disruption of PRC2 and enhancer functions.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All sequencing datasets generated during this work have been deposited in the Gene Expression Omnibus under accession number GSE154267. Source data are provided with this paper.
References
Mohammad, F. & Helin, K. Oncohistones: drivers of pediatric cancers. Genes Dev. 31, 2313–2324 (2017).
Jones, C. & Baker, S. J. Unique genetic and epigenetic mechanisms driving paediatric diffuse high-grade glioma. Nat. Rev. Cancer 14, 651–661 (2014).
Wu, G. et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat. Genet. 44, 251–253 (2012).
Schwartzentruber, J. et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 482, 226–231 (2012).
Goldberg, A. D. et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 140, 678–691 (2010).
Talbert, P. B. & Henikoff, S. Histone variants on the move: substrates for chromatin dynamics. Nat. Rev. Mol. Cell Biol. 18, 115–126 (2017).
Maze, I., Noh, K. M., Soshnev, A. A. & Allis, C. D. Every amino acid matters: essential contributions of histone variants to mammalian development and disease. Nat. Rev. Genet. 15, 259–271 (2014).
Mackay, A. et al. Integrated molecular meta-analysis of 1,000 pediatric high-grade and diffuse intrinsic pontine glioma. Cancer Cell 32, 520–537.e5 (2017).
Vinci, M. et al. Functional diversity and cooperativity between subclonal populations of pediatric glioblastoma and diffuse intrinsic pontine glioma cells. Nat. Med. 24, 1204–1215 (2018).
Nikbakht, H. et al. Spatial and temporal homogeneity of driver mutations in diffuse intrinsic pontine glioma. Nat. Commun. 7, 11185 (2016).
Pathania, M. et al. H3.3(K27M) cooperates with Trp53 loss and PDGFRA gain in mouse embryonic neural progenitor cells to induce invasive high-grade gliomas. Cancer Cell 32, 684–700.e9 (2017).
Larson, J. D. et al. Histone H3.3 K27M accelerates spontaneous brainstem glioma and drives restricted changes in bivalent gene expression. Cancer Cell 35, 140–155.e7 (2019).
Louis, D. N. et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 131, 803–820 (2016).
Weinberg, D. N., Allis, C. D. & Lu, C. Oncogenic mechanisms of histone H3 mutations. Cold Spring Harb. Perspect. Med. 7, a026443 (2017).
Lewis, P. W. et al. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science 340, 857–861 (2013).
Bracken, A. P., Brien, G. L. & Verrijzer, C. P. Dangerous liaisons: interplay between SWI/SNF, NuRD, and Polycomb in chromatin regulation and cancer. Genes Dev. 33, 936–959 (2019).
Conway, E., Healy, E. & Bracken, A. P. PRC2 mediated H3K27 methylations in cellular identity and cancer. Curr. Opin. Cell Biol. 37, 42–48 (2015).
Chan, K. M. et al. The histone H3.3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression. Genes Dev. 27, 985–990 (2013).
Bender, S. et al. Reduced H3K27me3 and DNA hypomethylation are major drivers of gene expression in K27M mutant pediatric high-grade gliomas. Cancer Cell 24, 660–672 (2013).
Justin, N. et al. Structural basis of oncogenic histone H3K27M inhibition of human Polycomb repressive complex 2. Nat. Commun. 7, 11316 (2016).
Diehl, K. L. et al. PRC2 engages a bivalent H3K27M-H3K27me3 dinucleosome inhibitor. Proc. Natl Acad. Sci. USA 116, 22152–22157 (2019).
Voigt, P. & Reinberg, D. Putting a halt on PRC2 in pediatric glioblastoma. Nat. Genet. 45, 587–589 (2013).
Jain, S. U. et al. H3 K27M and EZHIP impede H3K27-methylation spreading by inhibiting allosterically stimulated PRC2. Mol. Cell 80, 726–735.e7 (2020).
Harutyunyan, A. S. et al. H3K27M induces defective chromatin spread of PRC2-mediated repressive H3K27me2/me3 and is essential for glioma tumorigenesis. Nat. Commun. 10, 1262 (2019).
Mohammad, F. et al. EZH2 is a potential therapeutic target for H3K27M-mutant pediatric gliomas. Nat. Med. 23, 483–492 (2017).
Piunti, A. et al. Therapeutic targeting of Polycomb and BET bromodomain proteins in diffuse intrinsic pontine gliomas. Nat. Med. 23, 493–500 (2017).
Krug, B. et al. Pervasive H3K27 acetylation leads to ERV expression and a therapeutic vulnerability in H3K27M gliomas. Cancer Cell 35, 782–797.e8 (2019).
Sturm, D. et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell 22, 425–437 (2012).
Bressan, R. B. et al. Regional identity of human neural stem cells determines oncogenic responses to histone H3.3 mutants. Cell Stem Cell 28, 877–893.e9 (2021).
Bracken, A. P. et al. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev. 21, 525–530 (2007).
Cordero, F. J. et al. Histone H3.3K27M represses p16 to accelerate gliomagenesis in a murine model of DIPG. Mol. Cancer Res. 15, 1243–1254 (2017).
Banaszynski, L. A. et al. Hira-dependent histone H3.3 deposition facilitates PRC2 recruitment at developmental loci in ES cells. Cell 155, 107–120 (2013).
Fang, D. et al. H3.3K27M mutant proteins reprogram epigenome by sequestering the PRC2 complex to poised enhancers. eLife 7, e36696 (2018).
Rada-Iglesias, A. et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470, 279–283 (2011).
Ferrari, K. J. et al. Polycomb-dependent H3K27me1 and H3K27me2 regulate active transcription and enhancer fidelity. Mol. Cell 53, 49–62 (2014).
Nagaraja, S. et al. Histone variant and cell context determine H3K27M reprogramming of the enhancer landscape and oncogenic state. Mol. Cell 76, 965–980.e12 (2019).
Corces, M. R. et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods 14, 959–962 (2017).
Jessa, S. et al. Stalled developmental programs at the root of pediatric brain tumors. Nat. Genet. 51, 1702–1713 (2019).
Nagaraja, S. et al. Transcriptional dependencies in diffuse intrinsic pontine glioma. Cancer Cell 31, 635–652.e6 (2017).
Grasso, C. S. et al. Functionally defined therapeutic targets in diffuse intrinsic pontine glioma. Nat. Med. 21, 555–559 (2015).
Glancy, E., Ciferri, C. & Bracken, A. P. Structural basis for PRC2 engagement with chromatin. Curr. Opin. Struct. Biol. 67, 135–144 (2020).
Healy, E. et al. PRC2.1 and PRC2.2 synergize to coordinate H3K27 trimethylation. Mol. Cell 76, 437–452.e6 (2019).
Hojfeldt, J. W. et al. Non-core subunits of the PRC2 complex are collectively required for its target-site specificity. Mol. Cell 76, 423–436.e3 (2019).
Stafford, J. M. et al. Multiple modes of PRC2 inhibition elicit global chromatin alterations in H3K27M pediatric glioma. Sci. Adv. 4, eaau5935 (2018).
Zhang, T., Zhang, Z., Dong, Q., Xiong, J. & Zhu, B. Histone H3K27 acetylation is dispensable for enhancer activity in mouse embryonic stem cells. Genome Biol. 21, 45 (2020).
Conti, L. et al. Niche-independent symmetrical self-renewal of a mammalian tissue stem cell. PLoS Biol. 3, e283 (2005).
Bressan, R. B. et al. Efficient CRISPR/Cas9-assisted gene targeting enables rapid and precise genetic manipulation of mammalian neural stem cells. Development 144, 635–648 (2017).
Pollard, S. M. et al. Glioma stem cell lines expanded in adherent culture have tumor-specific phenotypes and are suitable for chemical and genetic screens. Cell Stem Cell 4, 568–580 (2009).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Orlando, D. A. et al. Quantitative ChIP-Seq normalization reveals global modulation of the epigenome. Cell Rep. 9, 1163–1170 (2014).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Ramirez, F. et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Lerdrup, M., Johansen, J. V., Agrawal-Singh, S. & Hansen, K. An interactive environment for agile analysis and visualization of ChIP-sequencing data. Nat. Struct. Mol. Biol. 23, 349–357 (2016).
Whyte, W. A. et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319 (2013).
Loven, J. et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153, 320–334 (2013).
Kim, D., Paggi, J. M., Park, C., Bennett, C. & Salzberg, S. L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915 (2019).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Ewels, P., Magnusson, M., Lundin, S. & Kaller, M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048 (2016).
Reske, J. J., Wilson, M. R. & Chandler, R. L. ATAC-seq normalization method can significantly affect differential accessibility analysis and interpretation. Epigenetics Chromatin 13, 22 (2020).
Lun, A. T. & Smyth, G. K. csaw: a Bioconductor package for differential binding analysis of ChIP-seq data using sliding windows. Nucleic Acids Res. 44, e45 (2016).
Ge, S. X., Jung, D. & Yao, R. ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics 36, 2628–2629 (2020).
Acknowledgements
We thank members of the A.P.B., S.M.P. and G.L.B. laboratories for helpful discussions and critical reading of the manuscript. We are grateful to the Genomics Core at University College Dublin for expertise and help with next-generation sequencing. Work in the A.P.B. laboratory is supported by Worldwide Cancer Research and The Brain Tumour Charity (18-0592), the Irish Research Council Advanced Laureate Award (IRCLA/2019/21), the Health Research Board (HRB-ILP-POR-2017-078), Science Foundation Ireland (SFI) under the SFI Investigators (SFI/16/IA/4562) and BBSRC-SFI (SFI/17/BBSRC/3415) programs, the Irish Cancer Society (CancersUnmetNeeds012) and the St. Vincent’s Foundation. O.D. was supported by a PhD fellowship from the Irish Research Council Government of Ireland Postgraduate Scholarship Programme (GOIPG/2017/2009). D.G. was supported by a PhD fellowship from the Irish Research Council Government of Ireland Postgraduate Scholarship Programme (GOIPG/2019/2084). R.B.B. was supported by a fellowship from the Science Without Borders Program (CAPES, Governo Dilma Rousseff, Brazil) and a postdoctoral fellowship from EMBO. S.M.P. was supported by a Cancer Research UK Senior Research Fellowship (A17368). S.M.P. and R.B.B. were supported by a project grant from Children with Cancer (15/189). Support for cellular models was provided by G. Morrison and the Cancer Research UK-funded Accelerator Award (A21922; http://gcgr.org.uk). Work in the G.L.B. laboratory is supported by an SFI Starting Investigator Research Grant (18/SIRG/5573), an Irish Cancer Society Biomedical Research Fellowship (CRF18BRI) and a Worldwide Cancer Research grant (21-0271).
Author information
Authors and Affiliations
Contributions
G.L.B., R.B.B., S.M.P. and A.P.B. conceived of the project and designed the experiments. R.B.B., V.G. and S.M.P. derived and engineered the human fetal hindbrain NSC cultures. G.L.B. performed most of the bench-based experiments, while R.B.B. and D.G. contributed to phenotypic characterizations of NSC cultures. E.L. performed EZH2 inhibitor treatment and global gene expression analyses in DIPGXIII cells. O.D. performed the ESC ChIP-RX analyses. R.B.B., M.-A.M.-T. and N.A. performed the xenotransplantation experiments. C.M., H.N., A.M.D., E.H. and D.J.F. performed the bioinformatics analyses. G.L.B., R.B.B., D.G. and A.P.B. co-wrote the manuscript with contributions from all other authors.
Corresponding authors
Ethics declarations
Competing interests
S.M.P. is a founder and shareholder of, and paid consultant to, Cellinta—a biotechnology start-up that is developing cancer therapeutics. S.M.P. is also an inventor on a University of Edinburgh patent related to NSC culture methods (WO2005121318A3). The other authors declare no competing interests.
Additional information
Peer review information Nature Genetics thanks Gerald Crabtree and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
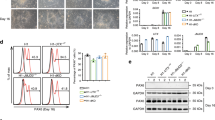
Extended Data Fig. 1 An isogenic hindbrain neural stem cell DMG model.
a. Immunoblots in cell lysates from unmanipulated NSCs and lines expressing WT or K27M-mutant H3.3. Representative example of n = 2 data. b. Absolute quantitative PCR of endogenous/exogenous H3F3A transcripts in NSCs expressing WT or K27M H3F3A, n = 3 independent samples. c. Immunoblots in cell lysates from WT or K27M H3.3 NSC cultures. Representative example of n = 2 data. d. Immunoblots (Left) and quantifications (Right) of total H3 levels in NSCs expressing WT or K27M H3F3A, n = 4 independent samples. Presented as mean values ± SD. e. Immunoblots in NSCs expressing WT or K27M H3F3A or H3K27M mutant DIPG cell lines. Representative example of n = 2 data. f. Relative quantitative PCR analysis of INK4A levels in NSCs expressing WT or K27M H3.3, n = 2 independent samples. g. Growth analysis of H3.3 WT and K27M expressing NSCs, n = 3 independent samples. Presented as mean values ± SD.
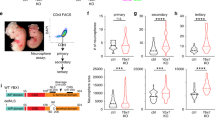
Extended Data Fig. 2 Tumorigenic potential of hindbrain NSC DMG model.
a. EdU positivity in PP5W and PP5K cultures derived from embryos GCGR-NS19 and GCGR-NS13, n = 4 independent samples. Presented as mean values ± SD. b. Colony forming activity in PP5W and PP5K cultures, n = 6 independent samples. Data are median values and interquartile ranges (IQR), with whiskers representing quartiles 1/3 ± (1.5 × IQR) respectively. c. Senescence Associated ß-Galactosidase (SA-ß-Gal) positivity in PP5W and PP5K cultures, n = 3 independent samples. Presented as mean values ± SD. d. Mouse survival following stereotactic injection of PP5W or PP5K NSCs, n = 4 PP5W and n = 8 PP5K mice. e. Brightfield and GFP micrographs of wholemount/cross-sectional views of a representative mouse brain injected with PP5K cells. f. Immunohistochemistry of brain samples from mice transplanted with PP5W and PP5K cells. Transplanted V5-tag (H3.3) positive cells display a diffuse pattern in mouse tissue, with no defined margins between human cells and the mouse brain parenchyma. Scale bar: 1.5mm. Representative example of n = 4 data. g. Immunohistochemistry of V5-tag (H3.3) and Ki67 in brain tissue from mice transplanted with PP5W and PP5K cells. Bar plot shows percentage of proliferating transplanted cells (KI67±V5-tag+) in each group. Scale bar: 20mM, n = 4 WT and n = 3 K27M biological samples. Presented as mean values ± SD.
Extended Data Fig. 3 Transcriptional and chromatin mapping in H3.3 WT and K27M expressing hindbrain NSCs.
a. RNA-seq volcano plot presenting the gene expression changes observed between NSC cultures expressing WT or K27M-mutant H3F3A. Red and blue denotes genes significantly up and down-regulated, respectively in K27M cultures. b. Box plots presenting the expression level of the indicated genes in a cohort of H3.3 WT (black) and K27M-mutant (red) DIPG patient samples. This data was downloaded from the pediatric cBioportal database (https://pedcbioportal.kidsfirstdrc.org), n = 118 WT and n = 71 H3.3 K27M independent tumour samples. The box plots present median values and interquartile ranges (IQR), with whiskers representing quartiles 1/3 ± (1.5 × IQR) respectively. c. Gene Set Enrichment Analysis (GSEA) of genes up- and down-regulated in K27M expressing NSC cultures compared to genes up- and down-regulated in K27M-mutant patient samples. d. Genome-wide correlations of H3.3 WT and K27M (V5) ChIP-seq read densities derived from embryo GCGR-NS13. The correlation coefficient for the two conditions is indicated. e. Violin plots representing the normalised abundance of H3.3 (V5) at the indicated genomic locations in embryos GCGR-NS19 and GCGR-NS13, in H3.3 WT (left panel) and K27M (right panel) conditions. An equivalent number of non-PRC2 target, repressed gene promoters (as compared to PRC2 target genes) were randomly selected from RNA-seq results, as an additional control set of genomic loci. Data presented are from each biologically independent embryo sample, n = 1/embryo. f. Meta-plots of average H3.3-WT ChIP-seq enrichment in biological duplicate experiments in genomic windows ±10kb of Active Promoter, Active Enhancer, PRC2 target promoters and non-PRC2 target Repressed gene promoters.
Extended Data Fig. 4 Increased SUZ12 chromatin binding in K27M-mutant hindbrain NSCs.
a. Venn diagrams presenting the total numbers and overlap of identified SUZ12 peak sets in WT and K27M-mutant NSC cultures (top panel). Meta-tracks presenting quantitative ChIP-Rx normalized SUZ12 signal at overlapping and distinct peak sets identified by Venn diagram analysis (middle panels). Meta-tracks presenting ChIP-seq normalized V5 H3.3 WT and K27M signal at the genomic regions outlined (bottom panels). b. Genomic tracks showing average SUZ12, H3K27me3 and BMI1 ChIP-Rx and H3.3-K27M ChIP-seq signal at the indicated genomic locus (chr6:125,737,994-125,772,907) in biological duplicate WT and K27M-mutant hindbrain NSC cultures. The chromosome ideograms are displayed above the gene track panels with the relevant regions highlighted. c. Box plots presenting the fold-change in quantitative SUZ12 ChIP-Rx signal at SUZ12 peaks and outside SUZ12 peaks between K27M-mutant and WT hindbrain NSC cultures for embryos GCGR-NS19 (left) and GCGR-NS13 (right). Data presented are from each biologically independent embryo sample, n = 1/embryo. Box plots present median values and interquartile ranges (IQR), with whiskers representing quartiles 1/3 ± (1.5 × IQR) respectively. d. Meta-tracks with density of CpG dinucleotides in a ± 10kb genomic window around the transcriptional start sites of genes gaining, with unchanged or reduced SUZ12 binding. e. Meta-tracks presenting ChIP-Rx normalised H3K4me3 levels at the promoter regions of the same genes as in panel b. f. Meta-tracks presenting ChIP-Rx (H3K27me3, H3K4me1, SUZ12, H3K4me3, H3K27ac) or ChIP-seq (H3.3A) normalised read counts at 263 identified poised enhancer elements in hindbrain NSC cultures.
Extended Data Fig. 5 PRC2 target gene dynamics in H3.3-K27M mutant NSCs.
a Gene ontology analysis of genes with unchanged (±0.1-fold) or reduced (>1.5-fold) SUZ12 binding in K27M-mutant NSC cultures. b Box plots presenting expression values for each of the indicated PRC2 target genes (Gaining SUZ12) in H3.3 WT or K27M mutant NSCs, n = 4 biologically independent samples. Box plots present median values and interquartile ranges (IQR), with whiskers representing quartiles 1/3 ± (1.5 × IQR) respectively.
Extended Data Fig. 6 H3K27 modification dynamics in K27M-mutant NSCs.
a. H3K27me3 peak number in WT and K27M NSC cultures. Total number of peaks in each condition are indicated, n = 2 independent samples. b. Number of ChIP-Rx normalised read counts for SUZ12 (left) and H3K27me3 (right) at ‘Targeted’ and ‘Dispersed’ regions in embryo GCGR-NS13. c. Fold-change in H3K27me3 ChIP-Rx signal at ‘Targeted’ and ‘Dispersed’ sites in WT and K27M hindbrain NSCs for embryos GCGR-NS19 (left) and GCGR-NS13 (right), n = 1/embryo. Data are median values and interquartile ranges (IQR), with whiskers representing quartiles 1/3 ± (1.5 × IQR) respectively. d. Tornado plots and meta tracks of ChIP-Rx normalised SUZ12 and H3K27me3 signal ±10kb of all SUZ12 peak regions in duplicate WT and K27M NSCs. e. Genomic tracks showing SUZ12, H3K27me2 and H3K27me3 ChIP-Rx signal at the indicated genomic loci (chr2:175,593,388-176,711,387 and chr7:155,162,247-156,015,446) in WT and K27M-mutant NSC cultures for embryos. f. Scatter density plots correlating change in H3K27me2 (left) and H3K27me3 (right) with H3K27ac in 10kb genomic bins between H3.3 WT and K27M NSCs. g. Rolling average plots presenting fold-change of H3K27 modifications in ChIP-Rx across chromosome 4 in Ezh2 wildtype and heterozygous knockout mouse ESCs. h. Genome-wide correlation of H3K27ac ChIP-Rx signal in WT and K27M conditions in embryo GCGR-NS13 (top panels) and Ezh2 wildtype and heterozygous knockout mouse ESCs (bottom panels). Correlations of H3K27ac ChIP-Rx read densities are also shown for low abundance regions (right panels). i. Violin plots presenting fold-change of H3K27 modifications in ChIP-Rx in 10kb bins grouped into quintiles based on H3K27ac abundance in WT cells (left panels). The number of bins in each quintile is indicated. Right panels show violin plots presenting the abundance H3K27 modifications within quintiles as per the left panels. Data presented are from each independent embryo sample, n = 1/embryo.
Extended Data Fig. 7 Enhancer dynamics in H3.3-K27M NSC cultures.
a. Ranked Order of Super Enhancer (ROSE) analysis in WT and K27M-mutant NSC cultures for embryo GCGR-NS13. b. Bar charts presenting the relative shift in the number of typical and super enhancer elements in WT and K27M-mutant hindbrain NSC cultures, n = 2 biologically independent samples. c. Tornado plots of averaged ChIP-Rx normalised H3K27ac signal ±10kb of the centre of active promoters and enhancer elements in duplicate WT and K27M-mutant NSC cultures. d. Meta-tracks of ChIP-Rx normalised signal for the indicated antibodies at active promoter and enhancer regions in duplicate WT and K27M-mutant NSC cultures. e. Waterfall plots presenting the log2 fold-change in H3K27ac ChIP-Rx signal at all identified active enhancers (top) and active promoters (bottom) between WT and K27M-mutant NSC cultures. f. Box plots presenting the log2 fold-change in H3K27ac ChIP-Rx signal at active promoters and enhancers separated into their respective top (Q5), and bottom (Q1) quintiles based on H3.3-K27M abundance, n = 2 biologically independent samples. Box plots present median values and interquartile ranges (IQR), with whiskers representing quartiles 1/3 ± (1.5 × IQR) respectively. g. Genomic tracks showing quantitative H3K27ac ChIP-Rx signal around the HOXA gene cluster on chromosome 7 (chr7:26,811,403-27,600,402). h. Meta-tracks of ChIP-seq normalised H3K27M signal at active promoter and enhancer regions in two independent K27M mutant patient derived DMG cell lines. One sample is H3.1-K27M, while the second is H3.3-K27M.
Extended Data Fig. 8 Gene ontology analysis of differentially expressed genes associated with gene enhancer dynamics.
a. Waterfall plots presenting the log2 fold-change in ATAC-seq signal at all identified active enhancers (left) and active promoters (right) between WT and K27M-mutant NSC cultures. b. Gene ontology (GO) analysis of all genes associated with enhancer regions losing both H3K27ac signal and chromatin accessibility in K27M mutant NSCs (left). GO analysis of downregulated genes associated with enhancer regions losing both H3K27ac signal and chromatin accessibility (right). c. GO analysis of all genes associated with enhancer regions losing H3K27ac signal only in K27M mutant NSCs (left). GO analysis of downregulated genes associated with enhancer regions losing H3K27ac (right).
Extended Data Fig. 9 Response of WT and K27M mutant NSCs to EZH2 inhibition.
a. Cellular viability dose-response in PP5W (black) or PP5K (red) cultures treated with the indicated small-molecules. Mean ± s.d., n = 3 independent samples. b. GO analysis of genes significantly upregulated (top panels) or downregulated (bottom panels) in PP5W and PP5K cultures following treatment with Tazemetostat. c. RNA-seq volcano plots showing expression changes in H3.3-K27M mutant mouse NSC cells treated with Tazemetostat (left) or a DMG cell line following shRNA SUZ12 knockdown (right). Total numbers of up/down regulated genes are indicated, as are the numbers of up/down regulated genes gaining SUZ12 at their promoters in H3.3-K27M human NSCs. In mouse cells, the homologs of genes gaining SUZ12 are highlighted. d. Plots showing log2 fold-change in expression for the set of non-PRC2 repressed loci in Tazemetostat treated PP5W or PP5K cultures, n = 3 independent samples. Presented are median values and interquartile ranges (IQR), with whiskers representing quartiles 1/3 ± (1.5 × IQR) respectively. e. Genomic tracks showing RNA-seq data at the INK4A-ARF locus (chr9:21,961,091-22,001,052) in PP5W and PP5K cultures treated with DMSO or Tazemetostat. Indicated are the INK4A and ARF specific exons for each transcript. f. Plots showing log2 expression values for the indicated genes in H3.3 WT or K27M NSCs (left panels), n = 4 independent samples or PP5K cells treated with DMSO or Tazemetostat (right panels), n = 3 independent samples. Presented are median values and interquartile ranges (IQR), with whiskers representing quartiles 1/3 ± (1.5 × IQR) respectively. g. Plots presenting the log2 fold-change in gene expression for the set of neurodevelopmental regulatory genes which lose H3K27ac and chromatin accessibility at their promoters in Tazemetostat treated PP5W or PP5K cultures, n = 3 independent samples. Presented are median values and interquartile ranges (IQR), with whiskers representing quartiles 1/3 ± (1.5 × IQR) respectively.
Extended Data Fig. 10 Transcriptional disruption in DIPG cell lines treated with compounds targeting chromatin regulators.
RNA-seq volcano plots presenting the gene expression changes observed in an established H3.3-K27M mutant DIPG cell, DIPGXIII following treatment with Tazemetostat, JQ1, THZ1 or Panobinostat for 24 hours. Red and blue denotes genes significantly up and down-regulated, respectively in drug treated cultures.
Supplementary information
Source data
Source Data Fig. 1
Unprocessed western blots and/or gels.
Source Data Extended Data Fig. 1
Unprocessed western blots and/or gels.
Rights and permissions
About this article
Cite this article
Brien, G.L., Bressan, R.B., Monger, C. et al. Simultaneous disruption of PRC2 and enhancer function underlies histone H3.3-K27M oncogenic activity in human hindbrain neural stem cells. Nat Genet 53, 1221–1232 (2021). https://doi.org/10.1038/s41588-021-00897-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-021-00897-w
This article is cited by
-
PRMT5 inhibition shows in vitro efficacy against H3K27M-altered diffuse midline glioma, but does not extend survival in vivo
Scientific Reports (2024)
-
Pediatric glioma histone H3.3 K27M/G34R mutations drive abnormalities in PML nuclear bodies
Genome Biology (2023)
-
Oncohistones and disrupted development in pediatric-type diffuse high-grade glioma
Cancer and Metastasis Reviews (2023)
-
Loss of p53 Concurrent with RAS and TERT Activation Induces Glioma Formation
Molecular Neurobiology (2023)
-
Oncohistone interactome profiling uncovers contrasting oncogenic mechanisms and identifies potential therapeutic targets in high grade glioma
Acta Neuropathologica (2022)