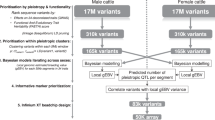
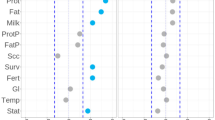
Abstract
Mammalian species carry ~100 loss-of-function variants per individual1,2, where ~1–5 of these impact essential genes and cause embryonic lethality or severe disease when homozygous3. The functions of the remainder are more difficult to resolve, although the assumption is that these variants impact fitness in less manifest ways. Here we report one of the largest sequence-resolution screens of cattle to date, targeting discovery and validation of non-additive effects in 130,725 animals. We highlight six novel recessive loci with impacts generally exceeding the largest-effect variants identified from additive genome-wide association studies, presenting analogs of human diseases and hitherto-unrecognized disorders. These loci present compelling missense (PLCD4, MTRF1 and DPF2), premature stop (MUS81) and splice-disrupting (GALNT2 and FGD4) mutations, together explaining substantial proportions of inbreeding depression. These results demonstrate that the frequency distribution of deleterious alleles segregating in selected species can afford sufficient power to directly map novel disorders, presenting selection opportunities to minimize the incidence of genetic disease.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
Genotype, phenotype and sequence datasets representing the different experimental populations have been deposited in the Dryad digital data repository (kwh70rz1z) and NCBI Sequence Read Archive (PRJNA656361, PRJNA682457 and PRJNA683948). Source data are provided with this paper.
Code availability
Scripts used in this study are available on GitHub. R scripts for power calculations can be found at https://github.com/egmreynolds/PowerCalculations.git. Julia source code for GWAS and pedigree-based association methods can be found at https://github.com/egmreynolds/MANA.git.
Change history
07 June 2021
In the version of this article initially published, the image of Extended Data Fig. 3 mistakenly duplicated that of Extended Data Fig. 6, and that of Extended Data Fig. 5 mistakenly duplicated that of Extended Data Fig. 8. The errors have been corrected in the HTML and PDF versions of the article.
References
Charlier, C. et al. NGS-based reverse genetic screen for common embryonic lethal mutations compromising fertility in livestock. Genome Res. 26, 1333–1341 (2016).
Lek, M. et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 (2016).
Gao, Z., Waggoner, D., Stephens, M., Ober, C. & Przeworski, M. An estimate of the average number of recessive lethal mutations carried by humans. Genetics 199, 1243–1254 (2015).
Daetwyler, H. D. et al. Whole-genome sequencing of 234 bulls facilitates mapping of monogenic and complex traits in cattle. Nat. Genet. 46, 858–865 (2014).
Littlejohn, M. D. et al. Functionally reciprocal mutations of the prolactin signalling pathway define hairy and slick cattle. Nat. Commun. 5, 5861 (2014).
Adams, H. A. et al. Identification of a nonsense mutation in APAF1 that is likely causal for a decrease in reproductive efficiency in Holstein dairy cattle. J. Dairy Sci. 99, 6693–6701 (2016).
Reed, D. R., Lawler, M. P. & Tordoff, M. G. Reduced body weight is a common effect of gene knockout in mice. BMC Genet. 9, 4 (2008).
Karim, L. et al. Variants modulating the expression of a chromosome domain encompassing PLAG1 influence bovine stature. Nat. Genet. 43, 405–413 (2011).
Cai, Z., Guldbrandtsen, B., Lund, M. S. & Sahana, G. Weighting sequence variants based on their annotation increases the power of genome-wide association studies in dairy cattle. Genet. Sel. Evol. 51, 20 (2019).
Finno, C. J. et al. A missense mutation in MYH1 is associated with susceptibility to immune-mediated myositis in Quarter Horses. Skelet. Muscle 8, 7 (2018).
Grisart, B. et al. Positional candidate cloning of a QTL in dairy cattle: identification of a missense mutation in the bovine DGAT1 gene with major effect on milk yield and composition. Genome Res. 12, 222–231 (2002).
Lopdell, T. J. et al. Multiple QTL underlie milk phenotypes at the CSF2RB locus. Genet. Sel. Evol. 51, 3 (2019).
Littlejohn, M. D. et al. Sequence-based association analysis reveals an MGST1 eQTL with pleiotropic effects on bovine milk composition. Sci. Rep. 6, 25376 (2016).
Vasileiou, G. et al. Mutations in the BAF-complex subunit DPF2 are associated with Coffin-Siris syndrome. Am. J. Hum. Genet. 102, 468–479 (2018).
Delague, V. et al. Mutations in FGD4 encoding the Rho GDP/GTP exchange factor FRABIN cause autosomal recessive Charcot-Marie-Tooth type 4H. Am. J. Hum. Genet. 81, 1–16 (2007).
Stendel, C. et al. Peripheral nerve demyelination caused by a mutant Rho GTPase guanine nucleotide exchange factor, Frabin/FGD4. Am. J. Hum. Genet. 81, 158–164 (2007).
Khetarpal, S. A. et al. Loss of function of GALNT2 lowers high-density lipoproteins in humans, nonhuman primates, and rodents. Cell Metab. 24, 234–245 (2016).
Zilmer, M. et al. Novel congenital disorder of O-linked glycosylation caused by GALNT2 loss of function. Brain 143, 1114–1126 (2020).
Ge, X., Wang, Y., Lam, K. S. & Xu, A. Metabolic actions of FGF21: molecular mechanisms and therapeutic implications. Acta Pharm. Sin. B 2, 350–357 (2012).
Buniello, A. et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 47, D1005–D1012 (2019).
Bolormaa, S. et al. Detailed phenotyping identifies genes with pleiotropic effects on body composition. BMC Genomics 17, 224 (2016).
Fukami, K. et al. Requirement of phospholipase Cδ4 for the zona pellucida-induced acrosome reaction. Science 292, 920–923 (2001).
Zhu, Z. et al. Dominance genetic variation contributes little to the missing heritability for human complex traits. Am. J. Hum. Genet. 96, 377–385 (2015).
Bolormaa, S. et al. Non-additive genetic variation in growth, carcass and fertility traits of beef cattle. Genet. Sel. Evol. 47, 26 (2015).
Jiang, J. et al. Dissection of additive, dominance, and imprinting effects for production and reproduction traits in Holstein cattle. BMC Genomics 18, 425 (2017).
Jiang, J. et al. A large-scale genome-wide association study in U.S. Holstein cattle. Front. Genet. 10, 412 (2019).
Pausch, H. et al. Evaluation of the accuracy of imputed sequence variant genotypes and their utility for causal variant detection in cattle. Genet. Sel. Evol. 49, 24 (2017).
Advisory Committee on Traits Other than Production. Evaluation System for Traits Other than Production (TOP) for Dairy Cattle in New Zealand (DairyNZ, 2014).
Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at https://arxiv.org/abs/1303.3997 (2013).
DePristo, M. A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498 (2011).
Liao, Y., Smyth, G. K. & Shi, W. FeatureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Fink, T. et al. A new mechanism for a familiar mutation – bovine DGAT1 K232A modulates gene expression through multi-junction exon splice enhancement. BMC Genomics 21, 591 (2020).
Lucy, M. C. et al. Somatotropic axis components and nutrient partitioning in genetically diverse dairy cows managed under different feed allowances in a pasture system. J. Dairy Sci. 92, 526–539 (2009).
Browning, B. L. & Browning, S. R. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am. J. Hum. Genet. 84, 210–223 (2009).
Jivanji, S. et al. Genome-wide association analysis reveals QTL and candidate mutations involved in white spotting in cattle. Genet. Sel. Evol. 51, 62 (2019).
Fernando, R. L. & Garrick, D. in Genome-Wide Association Studies and Genomic Prediction (eds Gondro, C. et al.) 237–274 (Humana, 2013).
Bernal Rubio, Y. L. et al. Meta-analysis of genome-wide association from genomic prediction models. Anim. Genet. 47, 36–48 (2016).
Yang, J. et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet. 44, 369–375 (2012).
Cingolani, P. et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6, 80–92 (2012).
Sargolzaei, M. & Schenkel, F. S. QMSim: a large-scale genome simulator for livestock. Bioinformatics 25, 680–681 (2009).
Brito, F. V., Neto, J. B., Sargolzaei, M., Cobuci, J. A. & Schenkel, F. S. Accuracy of genomic selection in simulated populations mimicking the extent of linkage disequilibrium in beef cattle. BMC Genet. 12, 80 (2011).
Acknowledgements
We acknowledge the contributions of T. Hale, S. Delaney, L. Falleni and E. Barnaby at AgResearch Ruakura for management of animals during the farm trial experiments. We also thank K. Brown and A. Yeates of the Livestock Improvement Corporation, S. Morgan and the Farm Technical Team of DairyNZ, F. Hoekstra of VeeHof, K. Schutz and F. Huddart of AgResearch, and C. Rogers, B. Adams, K. Legg, R. Holleboom, B. Ilton and J. Huxley of Massey University for experimental advice and helping to perform these investigations. We also acknowledge J.-M. Wit of the Leiden University Medical Centre for facilitating discussions and contributing ideas regarding the GALNT2 work. We are also particularly grateful for financial support from the Ministry of Business, Innovation and Employment (MBIE; Wellington, New Zealand) and the Ministry for Primary Industries (MPI; Wellington, New Zealand), who independently co-funded the work through the Endeavour Fund (LICX1802) and (now historical) Primary Growth Partnership research programs, respectively. E.G.M.R. is also supported by a Ministry for Primary Industries Postgraduate Scholarship (Wellington, New Zealand) and an Al Rae Centre scholarship (Hamilton, New Zealand). We are additionally grateful for the use of New Zealand eScience Infrastructure (NeSI) high-performance computing facilities as part of this research.
Author information
Authors and Affiliations
Contributions
Jointly supervised research: S.R.D., B.H., R.J.S., D.J.G., M.D.L. Conceived and designed the experiments: E.G.M.R., C.N., K.D., S.R.D., M.G., D.J.G., M.D.L. Performed the experiments: E.G.M.R., C.N., T.L., K.D., C.H., G.W., M.W., S.R.D., M.G., M.D.L. Performed statistical analysis: E.G.M.R., T.L., M.K., R.S., M.G., M.D.L. Analyzed the data: E.G.M.R., C.N., T.L., M.K., K.D., C.H., C. Couldrey, K.C., M.D.L. Contributed reagents/materials/analysis tools: E.G.M.R., T.L., K.D., C. Couldrey, T.J., K.T., M.W., R.S., K.C., B.H., C. Charlier, M.G., R.J.S., D.J.G. Wrote the paper: E.G.M.R., C.N., T.L., D.J.G., M.D.L.
Corresponding author
Ethics declarations
Competing interests
C.N., T.L., C.H., C. Couldrey, T.J., K.T., G.W., M.W., S.R.D., R.S., K.C., B.H., R.J.S. and M.D.L. are paid employees of the Livestock Improvement Corporation, a breeding company and supplier of bovine germplasm. The Livestock Improvement Corporation is also the applicant for several patent applications related to some of the mutations detailed in this article, with E.G.M.R., T.L., M.K., C.H., K.T., R.J.S. and M.D.L. named inventors on these applications. Specifically, these patents relate to genetic testing applications of mutations impacting the PLCD4 (749566; filed), GALNT2 (625688; granted), DPF2 (751917; filed), MUS81 (768801; filed), FGD4 (749555; filed), MTRF1 (751919; filed) and MYH1 (756281; filed) genes. All other authors declare no competing interests.
Additional information
Peer review information Nature Genetics thanks Aurelien Capitan, Ben Hayes and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Power calculations for different association models and sample sizes.
Plot contrasting the power of detection for different phenotype sources (i.e. cows, sires), different sample sizes, and different models (additive, recessive, class effect, and additive – without affected sires) given a locus explaining 0.1% of the phenotypic variance with a minor allele frequency of 2.5%, and heritability of 0.25. For sire models, each genotyped sire is assumed to have 100 un-genotyped daughters. Note that although sire models generally present higher power than cow models for a given number of genotyped animals, these models are not directly comparable since analyses based on breeding values typically leverage far fewer sires than studies using cows directly. Also note power for these sire models is provided for context only, since breeding values were not leveraged in analyses reported in this manuscript.
Extended Data Fig. 2 Dominance and additive QTL contrasts of allele frequencies, effect sizes, genotype group means, and p-values.
a, Plot contrasting minor allele frequency (MAF) and absolute effect size (Effect size, kg) of QTL identified in the standard-additive model (blue), and the dominance model (red) for body weight. Note that for equitable effect size comparison both additive and dominance estimates are represented as allele substitutions (i.e. effect of the heterozygote compared to the reference homozygote), so dominance effects only represent half the effect observed in homozygous individuals. b, Chromosome-wide scatterplots contrasting P-values of the eight recessive QTL for the standard-additive model (p_standard_additive) and dominance model (p_additive/p_dominance). P-values were computed using Z-tests, the genome-wide significance thresholds of p = 5 × 10−8 are drawn in red, an x = y line is drawn in black. Note that only seven chromosomes are presented since two effects were identified on chromosome 2 (and thus are not readily differentiated). c, Box plots showing adjusted-bodyweight genotype means for the 8 non-additive loci in the discovery population (N=79,945 cows). Genotypes used for display represent putative causative mutations or lead associated variants where no obvious candidate was identified. Note the largely additive effect presented by the PLAG1 locus, highlighted in GWAS due to a partial dominance effect. Box plots show median (centre line), interquartile range (box limits), and upper and lower whiskers (maxima and minima data points).
Extended Data Fig. 3 Manhattan plot of Chr19 recessive QTL showing MYH1 frameshifting indel.
a, 1.5Mbp sequence interval showing the top genome-wide non-additive association signal from analysis of body condition score in 75,617 cows; P-values were calculated using Z-tests. The genome-wide significance threshold of P < 5 × 10−8 is indicated by the horizontal grey line, note no variants at this locus surpassed this threshold (smallest P=7.6 × 10−8). Lead variants of the signal tag a 78 bp compound insertion deletion variant evident from inspection of whole genome sequence alignments. b, Genome sequence alignment of homozygous animal shown, resulting in predicted knockout of MYH1 due to simultaneous loss of 19 amino acids and introduction of a premature stop codon at exon 34.
Extended Data Fig. 4 Per-junction and per-gene Manhattan plots for splicing efficiency and gene expression QTL.
Manhattan plots showing splicing efficiency and whole-transcript expression QTL effects (P-values calculated using 2-sided t-tests). Splicing efficiency analysis was performed for the FGD4 (a) and GALNT2 (b) genes, with associations highlighting junctions for which splicing appears to be genetically modulated in cis. The proportion of spliced to un-spliced reads at each junction has been treated as an individual phenotype, with association analysis performed using intervals of imputed sequence data spanning the annotated gene structures, and ± 100kbp 5’ and 3’ of the gene boundaries. The splicing junction predicted to be impacted by the splice donor (FGD4) and acceptor (GALNT2) mutations is indicated by the blue highlighted panels, with the candidate causative mutation indicated in red. Whole transcript eQTL analysis was performed to assess possible gene expression impacts as a consequence of non-sense mediated RNA decay (NMD) for the FGD4 (c), GALNT2 (d), and MUS81 (e) genes that harbour non-sense mutations. Note that for the two genes that show significant eQTL (FGD4 and MUS81), the mutant allele is overexpressed and thus no NMD is apparent. In the case of the MUS81 and GALNT2 genes, lack of apparent NMD can be anticipated given the position of the GALNT2 c.1561-1G>A mutation in the final exon, and the presence of an in-frame start codon (p.Met76) following the p.Gly70 mutation for MUS81.
Extended Data Fig. 5 Splicing consequences of FGD4 and GALNT2 essential splice mutations.
Mammary RNA-seq alignments for the FGD4 (a) and GALNT2 (b) genes, showing wildtype and carrier animals for the FGD4 c.1671+1G>A and GALNT2 c.1561-1G>A splice mutations (two animals representing each genotype class per gene). Intron and exon numbers reference the ENSBTAT00000007175.5 and ENSBTAT00000006404.5 transcript annotations for the FGD4 and GALNT2 genes respectively. Right-most panels show intron-exon boundaries of the mutation-implicated splice junction, left-most panels show kilobase-level views of the whole intron and adjoining exon junction. Coverage tracks demonstrate clear intron retention for FGD4 heterozygous mutants, without obvious cryptic splicing. Animals heterozygous for GALNT2 mutant transcripts show less uniform intron retention, though at least three recurrent cryptic splice sites indicated by the purple arrows (green arrows show annotated junctions). c Putative translations for these alternatively spliced transcripts are indicated (light blue=reference splice, red=mis-splice), where the first base of the new acceptor exon boundaries are: cryptic 1 chr28 g.1309085; cryptic 2 chr28 g.1312087; cryptic 3 chr28 g.1312203. Note that all intron retention and cryptic splices are predicted to cause premature termination, with the exception of the ‘GALNT2 cryptic 3’ isoform that encodes a 44aa 5’ frame-extension of exon 16.
Extended Data Fig. 6 Hoof anatomical observations for DPF2 mutant and control individuals.
Figure showing hoof characteristics of DPF2 mutant and control cattle. Photographs (a), show the right rear hooves of representative mutants and controls, with the hooves of some mutants showing subjective differences including overlapping claw-tips and longer claws overall compared to controls (see centre two animals in mutant group). However, quantitative comparisons based on hoof measurements (b) did not reveal significant differences between groups (N=8 mutant and 9 control animals respectively; Supplementary Table 8). Box plots show median (centre line), interquartile range (box limits), and upper and lower whiskers (maxima and minima data points).
Extended Data Fig. 7 Nerve histology of FGD4 mutant and control individuals.
Common digital nerve of forelimb from two different FGD4 homozygotes (a and b) showing hypercellularity, Schwann cell hyperplasia, axonal swelling and degeneration (black arrow) (2000X, HE). c, Common digital nerve of forelimb from control animal (2000X, HE). d, Saphenous nerve from an FGD4 homozygote showing lack of myelin staining consistent with demyelination (2000X, Luxol fast blue). e, Saphenous nerve from a control animal (2000X, Luxol fast blue). Micrographs are representative of the lesions found in 7 different nerves examined from 2 FGD4 homozygotes and 2 control animals. Each nerve was examined in 3 locations, with both transverse and longitudinal sections. Bar = 50 µm.
Extended Data Fig. 8 Photographs of GALNT2 mutant and control individuals; PLCD4 mutant and control individuals.
Photographs contrasting homozygous mutant and homozygous reference animals for the GALNT2 c.1561-1G>A splice acceptor mutation (a), and PLCD4 p.Ala326Thr mutation (b). For the PLCD4 variant, front and rear images contrast the same two animals. Animals represent individuals from the research farm studies that were neither the smallest nor largest animals within each of their genotype classes. Photos are unstandardised and provided for qualitative purposes.
Extended Data Fig. 9 GWAS Manhattan plots exploring sensitivity of QTL detection for reduced resolution genotype data, and simulated loci varying in effect size and frequency.
Manhattan plot showing impact of marker density on discovery of non-additive bodyweight GWAS signals (a; P-values computed using Z-tests, horizontal grey line indicates the genome-wide significance threshold of P < 5 × 10−8). Here, dominance estimates from sequence-based bodyweight GWAS (grey dots) are plotted alongside a subsetted version of these same data filtered to represent the content of BovineSNP50k SNP-chip platform (green dots). While two of the modest effect, comparatively higher MAF QTL retain significance (i.e. Chr2:22Mbp and PLAG1 locus), only the DPF2/MUS81 QTL is represented among the major-effect, recessive signals. (b) Manhattan plot showing the influence of MAF and effect size on sensitivity of detection in a simulated dataset. Dominance estimates (blue dots) are contrasted with standard-additive estimates (grey dots), showing sensitivity of detection for 30 recessive causative mutations (red dots). Recessive effects were generated by randomly selecting variants from 1-5% MAF bins from the pool of simulated genotypes (frequencies indicated at bottom), with effect sizes assigned as 0.5 standard deviations (SD; light orange) or 1.0 SD (dark orange) per mutation. Mutations were selected to represent all chromosomes (two on chromosome 1).
Extended Data Fig. 10 Visualisation of sequence imputation allelic R-squared statistics by minor allele frequency, dominance effect sizes, and dominance p-values in the GWAS dataset.
Plots showing imputation allelic R2 (AR2) values of genotypes from the discovery population, where AR2 is taken to reflect accuracy of imputation, representing the squared correlation between the allele dosage with the highest posterior probability and the true allele dosage35. a, Box plots showing distributions of AR2 within different MAF classes for the 16,128,757 sequence variants used for GWAS. Box plots show median (centre line), interquartile range (box limits), and upper and lower whiskers (maxima and minima data points). b, Plot showing absolute dominance effect size (Effect size, kg) for genome-wide significant variants (P < 5 × 10−8) from the bodyweight GWAS, visualised by AR2. Also indicated are the candidate causative mutations of interest; effects are expressed as allele substitutions and thus represent half the effect observed in homozygous mutant individuals c, Scatter/density plot showing relationship between P-value and AR2 for the sequence variants tested in the bodyweight GWAS (dominance model), with mutations of interest also indicated.
Supplementary information
Source data
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Rights and permissions
About this article
Cite this article
Reynolds, E.G.M., Neeley, C., Lopdell, T.J. et al. Non-additive association analysis using proxy phenotypes identifies novel cattle syndromes. Nat Genet 53, 949–954 (2021). https://doi.org/10.1038/s41588-021-00872-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-021-00872-5
This article is cited by
-
X-linked genes influence various complex traits in dairy cattle
BMC Genomics (2023)
-
Dominance is common in mammals and is associated with trans-acting gene expression and alternative splicing
Genome Biology (2023)
-
Joint analysis of phenotype-effect-generation identifies loci associated with grain quality traits in rice hybrids
Nature Communications (2023)
-
Tutorial: a statistical genetics guide to identifying HLA alleles driving complex disease
Nature Protocols (2023)
-
A 1-bp deletion in bovine QRICH2 causes low sperm count and immotile sperm with multiple morphological abnormalities
Genetics Selection Evolution (2022)