Abstract
ATP-dependent chromatin remodelers are commonly mutated in human cancer. Mammalian SWI/SNF complexes comprise three conserved multisubunit chromatin remodelers (cBAF, ncBAF and PBAF) that share the BRG1 (also known as SMARCA4) subunit responsible for the main ATPase activity. BRG1 is the most frequently mutated Snf2-like ATPase in cancer. In the present study, we have investigated the role of SWI/SNF in genome instability, a hallmark of cancer cells, given its role in transcription, DNA replication and DNA-damage repair. We show that depletion of BRG1 increases R-loops and R-loop-dependent DNA breaks, as well as transcription–replication (T-R) conflicts. BRG1 colocalizes with R-loops and replication fork blocks, as determined by FANCD2 foci, with BRG1 depletion being epistatic to FANCD2 silencing. Our study, extended to other components of SWI/SNF, uncovers a key role of the SWI/SNF complex, in particular cBAF, in helping resolve R-loop-mediated T-R conflicts, thus, unveiling a new mechanism by which chromatin remodeling protects genome integrity.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The siBRG1 DRIPc-seq and RNA-seq datasets, the first siC DRIPc-seq replicate and one DRIP-seq replicate dataset have been deposited at the Gene Expression Omnibus repository and are available under accession no. GSE154631. Original data for another DRIP-seq replicate, siC RNA-seq and the second siC DRIPc-seq replicate dataset are available at the same database under accession no. GSE127979 (ref. 14), even though all the experiments were performed in parallel. Other publicly available genome-wide data used in the present study are listed in Supplementary Table 1. Cancer-related information for SWI/SNF genes was retrieved from cBio Cancer Genomics Portal (www.cbioportal.org)35,107 and Integrative Onco Genomics (www.intogen.org)108,109 databases. Source data are provided with this paper.
Code availability
Software and algorithms source and links are listed in Supplementary Table 1.
References
Gómez-González, B. & Aguilera, A. Transcription-mediated replication hindrance: a major driver of genome instability. Genes Dev. 33, 1008–1026 (2019).
García-Muse, T. & Aguilera, A. R loops: from physiological to pathological roles. Cell 179, 604–618 (2019).
Crossley, M. P., Bocek, M. & Cimprich, K. A. R-loops as cellular regulators and genomic threats. Mol. Cell 73, 398–411 (2019).
Domínguez-Sánchez, M. S., Barroso, S., Gómez-González, B., Luna, R. & Aguilera, A. Genome instability and transcription elongation impairment in human cells depleted of THO/TREX. PLoS Genet. 7, e1002386 (2011).
Huertas, P. & Aguilera, A. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol. Cell 12, 711–721 (2003).
Li, X. & Manley, J. L. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell 122, 365–378 (2005).
Skourti-Stathaki, K., Proudfoot, N. J. & Gromak, N. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol. Cell 42, 794–805 (2011).
Paulsen, R. D. et al. A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol. Cell 35, 228–239 (2009).
Wahba, L., Amon, J. D., Koshland, D. & Vuica-Ross, M. RNase H and multiple RNA biogenesis factors cooperate to prevent RNA:DNA hybrids from generating genome instability. Mol. Cell 44, 978–988 (2011).
Tuduri, S. et al. Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat. Cell Biol. 11, 1315–1324 (2009).
Aguilera, A. & Gómez-González, B. DNA–RNA hybrids: the risks of DNA breakage during transcription. Nat. Struct. Mol. Biol. 24, 439–443 (2017).
Cerritelli, S. M., Crouch, R. J. & Ribonuclease, H. The enzymes in eukaryotes. FEBS J. 276, 1494–1505 (2009).
Sreerama Chaitanya Sridhara, A., Carvalho, S., Rita Grosso, A., Marcela Gallego-Paez, L. & Carmo-Fonseca, M. Transcription dynamics prevent RNA-mediated genomic instability through SRPK2-dependent DDX23 phosphorylation. Cell Rep. 18, 334–343 (2017).
Pérez-Calero, C. et al. UAP56/DDX39B is a major cotranscriptional RNA–DNA helicase that unwinds harmful R loops genome-wide. Genes Dev. 34, 898–912 (2020).
Hatchi, E., Skourti-Stathaki, K., Proudfoot, N. J. & Livingston Correspondence, D. M. BRCA1 recruitment to transcriptional pause sites is required for R-loop-driven DNA damage repair. Mol. Cell 57, 636–647 (2015).
Bhatia, V. et al. BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature 511, 362–365 (2014).
García-Rubio, M. L. et al. The Fanconi anemia pathway protects genome integrity from R-loops. PLoS Genet. 11, e1005674 (2015).
Schwab, R. A. et al. The Fanconi anemia pathway maintains genome stability by coordinating replication and transcription. Mol. Cell 60, 351–361 (2015).
Herrera-Moyano, E., Mergui, X., García-Rubio, M. L., Barroso, S. & Aguilera, A. The yeast and human FACT chromatin reorganizing complexes solve R-loop mediated transcription-replication conflicts. Genes Dev. 28, 735–748 (2014).
Salas-Armenteros, I. et al. Human THO–Sin3A interaction reveals new mechanisms to prevent R-loops that cause genome instability. EMBO J. 36, 3532–3547 (2017).
Poli, J. et al. Mec1, INO80, and the PAF1 complex cooperate to limit transcription replication conflicts through RNAPII removal during replication stress. Genes Dev. 30, 337–354 (2016).
Feldman, J. L. & Peterson, C. L. Yeast sirtuin family members maintain transcription homeostasis to ensure genome stability. Cell Rep. 27, 2978–2989.e5 (2019).
Bayona-Feliu, A., Casas-Lamesa, A., Reina, O., Bernués, J. & Azorín, F. Linker histone H1 prevents R-loop accumulation and genome instability in heterochromatin. Nat. Commun. 8, 283 (2017).
Almeida, R. et al. Chromatin conformation regulates the coordination between DNA replication and transcription. Nat. Commun. 9, 1590 (2018).
Nguyen, D. T. et al. The chromatin remodelling factor ATRX suppresses R‐loops in transcribed telomeric repeats. EMBO Rep. 18, 914–928 (2017).
Chen, P. B., Chen, H. V., Acharya, D., Rando, O. J. & Fazzio, T. G. R loops regulate promoter-proximal chromatin architecture and cellular differentiation. Nat. Struct. Mol. Biol. 22, 999–1007 (2015).
Hodges, C., Kirkland, J. G. & Crabtree, G. R. The many roles of BAF (mSWI/SNF) and PBAF complexes in cancer. Cold Spring Harb. Perspect. Med. 6, a026930 (2016).
Kadoch, C. et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat. Genet. 45, 592–601 (2013).
Neigeborn, L. & Carlson, M. Genes affecting the regulation of SUC2 gene expression by glucose repression in Saccharomyces cerevisiae. Genetics 108, 845–858 (1984).
Stern, M., Jensen, R. & Herskowitz, I. Five SWI genes are required for expression of the HO gene in yeast. J. Mol. Biol. 178, 853–868 (1984).
Wilson, B. G. & Roberts, C. W. M. SWI/SNF nucleosome remodellers and cancer. Nat. Rev. Cancer 11, 481–492 (2011).
Centore, R. C., Sandoval, G. J., Soares, L. M. M., Kadoch, C. & Chan, H. M. Mammalian SWI/SNF chromatin remodeling complexes: emerging mechanisms and therapeutic strategies. Trends Genet. 36, 936–950 (2020).
Phelan, M. L., Sif, S., Narlikar, G. J. & Kingston, R. E. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol. Cell 3, 247–253 (1999).
Euskirchen, G. M. et al. Diverse roles and interactions of the SWI/SNF chromatin remodeling complex revealed using global approaches. PLoS Genet. 7, e1002008 (2011).
Cerami, E. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012).
Dykhuizen, E. C. et al. BAF complexes facilitate decatenation of DNA by topoisomerase IIα. Nature 497, 624–627 (2013).
Schlacher, K., Wu, H. & Jasin, M. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell 22, 106–116 (2012).
Pichierri, P., Franchitto, A. & Rosselli, F. BLM and the FANC proteins collaborate in a common pathway in response to stalled replication forks. EMBO J. 23, 3154–3163 (2004).
Zhou, B. et al. Comprehensive, integrated, and phased whole-genome analysis of the primary ENCODE cell line K562. Genome Res. 29, 472–484 (2019).
Sanz, L. A. et al. Prevalent, dynamic, and conserved r-loop structures associate with specific epigenomic signatures in mammals. Mol. Cell 63, 167–178 (2016).
Dunham, I. et al. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Mailand, N., Gibbs-Seymour, I. & Bekker-Jensen, S. Regulation of PCNA–protein interactions for genome stability. Nat. Rev. Mol. Cell Biol. 14, 269–282 (2013).
Zaborowska, J., Egloff, S. & Murphy, S. The pol II CTD: new twists in the tail. Nat. Struct. Mol. Biol. 23, 771–777 (2016).
Ashley, A. K. et al. DNA-PK phosphorylation of RPA32 Ser4/Ser8 regulates replication stress checkpoint activation, fork restart, homologous recombination and mitotic catastrophe. DNA Repair (Amst.) 121, 131–139 (2014).
Ward, I. M. & Chen, J. Histone H2AX Is phosphorylated in an ATR-dependent manner in response to replicational stress. J. Biol. Chem. 276, 47759–47762 (2001).
Wu, X. et al. Developmental and cancer-associated plasticity of DNA replication preferentially targets GC-poor, lowly expressed and late-replicating regions. Nucleic Acids Res. 46, 10157–10172 (2018).
Richardson, C. D. et al. CRISPR–Cas9 genome editing in human cells occurs via the Fanconi anemia pathway. Nat. Genet. 50, 1132–1139 (2018).
Pang, B., De Jong, J., Qiao, X., Wessels, L. F. A. & Neefjes, J. Chemical profiling of the genome with anti-cancer drugs defines target specificities. Nat. Chem. Biol. 11, 472–480 (2015).
Schick, S. et al. Systematic characterization of BAF mutations provides insights into intracomplex synthetic lethalities in human cancers. Nat. Genet. 51, 1399–1410 (2019).
Barisic, D., Stadler, M. B., Iurlaro, M. & Schübeler, D. Mammalian ISWI and SWI/SNF selectively mediate binding of distinct transcription factors. Nature 569, 136–140 (2019).
Pan, J. et al. The ATPase module of mammalian SWI/SNF family complexes mediates subcomplex identity and catalytic activity-independent genomic targeting. Nat. Genet. 51, 618–626 (2019).
Hu, G. et al. Regulation of nucleosome landscape and transcription factor targeting at tissue-specific enhancers by BRG1. Genome Res. 21, 1650–1658 (2011).
Dunaief, J. L. et al. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell 79, 119–130 (1994).
Cohen, S. M. et al. BRG1 co-localizes with DNA replication factors and is required for efficient replication fork progression. Nucleic Acids Res. 38, 6906–6919 (2010).
Wang, H. et al. BRCA1/FANCD2/BRG1-driven DNA repair stabilizes the differentiation state of human mammary epithelial cells. Mol. Cell https://doi.org/10.1016/j.molcel.2016.05.038 (2016).
Bochar, D. A. et al. BRCA1 is associated with a human SWI/SNF-related complex: linking chromatin remodeling to breast cancer. Cell 102, 257–265 (2000).
Park, J. H. et al. Mammalian SWI/SNF complexes facilitate DNA double-strand break repair by promoting γ-H2AX induction. EMBO J. 25, 3986–3997 (2006).
Lee, H. S., Park, J. H., Kim, S. J., Kwon, S. J. & Kwon, J. A cooperative activation loop among SWI/SNF, γ-H2AX and H3 acetylation for DNA double-strand break repair. EMBO J. 29, 1434–1445 (2010).
Sollier, J. et al. Transcription-coupled nucleotide excision repair factors promote r-loop-induced genome instability. Mol. Cell 56, 777–785 (2014).
Barroso, S. et al. The DNA damage response acts as a safeguard against harmful DNA–RNA hybrids of different origins. EMBO Rep. 20, e47250 (2019).
Yanling Zhao, D. et al. SMN and symmetric arginine dimethylation of RNA polymerase II C-terminal domain control termination. Nature 529, 48–53 (2016).
Alzu, A. et al. Senataxin associates with replication forks to protect fork integrity across RNA-polymerase-II-transcribed genes. Cell 151, 835–846 (2012).
Brambati, A. et al. Dormant origins and fork protection mechanisms rescue sister forks arrested by transcription. Nucleic Acids Res. 46, 122–1239 (2018).
Mischo, H. E. et al. Cell-cycle modulation of transcription termination factor Sen1. Mol. Cell 170, 312–326 (2018).
Otsuki, T. et al. Fanconi anemia protein, FANCA, associates with BRG1, a component of the human SWI/SNF complex. Hum. Mol. Genet. 10, 2651–2660 (2001).
Cristini, A., Groh, M., Kristiansen, M. S. & Gromak, N. RNA/DNA hybrid interactome identifies DXH9 as a molecular player in transcriptional termination and R-loop-associated DNA damage. Cell Rep. 23, 1891–1905 (2018).
Prado, F. & Aguilera, A. Impairment of replication fork progression mediates RNA polII transcription-associated recombination. EMBO J. 24, 1267–1276 (2005).
Hamperl, S., Bocek, M. J., Saldivar, J. C., Swigut, T. & Cimprich, K. A. Transcription–replication conflict orientation modulates R-loop levels and activates distinct dna damage responses. Cell 170, 774–786 (2017).
Lang, K. S. et al. Replication–transcription conflicts generate R-loops that orchestrate bacterial stress survival and pathogenesis. Cell 170, 787–799.e18 (2017).
García-Rubio, M. et al. Yra1-bound RNA–DNA hybrids cause orientation-independent transcription–replication collisions and telomere instability. Genes Dev. 32, 965–977 (2018).
Achar, Y. J., Adhil, M., Choudhary, R., Gilbert, N. & Foiani, M. Negative supercoil at gene boundaries modulates gene topology. Nature 577, 701–705 (2020).
Reyes, J. C. et al. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2α). EMBO J. 17, 6878–6991 (1998).
Muchardt, C., Bourachot, B., Reyes, J. C. & Yaniv, M. ras transformation is associated with decreased expression of the brm/SNF2α ATPase from the mammalian SWI–SNF complex. EMBO J. 17, 223–231 (1998).
Watanabe, R. et al. SWI/SNF factors required for cellular resistance to dna damage include arid1a and arid1b and show interdependent protein stability. Cancer Res. 74, 2465–2475 (2014).
Niimi, A., Chambers, A. L., Downs, J. A. & Lehmann, A. R. A role for chromatin remodellers in replication of damaged DNA. Nucleic Acids Res. 40, 393–403 (2012).
Kakarougkas, A. et al. Requirement for PBAF in transcriptional repression and repair at DNA beaks in actively transcribed regions of chromatin. Mol. Cell 55, 723–732 (2014).
Meisenberg, C. et al. Repression of transcription at DNA breaks requires cohesin throughout interphase and prevents genome instability. Mol. Cell 73, 212–223.e7 (2019).
Nielsen, A. L. et al. Selective interaction between the chromatin-remodeling factor BRG1 and the heterochromatin-associated protein HP1α. EMBO J. 21, 5797–5806 (2002).
Sif, S., Saurin, A. J., Imbalzano, A. N. & Kingston, R. E. Purification and characterization of mSin3A-containing Brg1 and hBrm chromatin remodeling complexes. Genes Dev. 15, 603–618 (2001).
Halazonetis, T. D., Gorgoulis, V. G. & Bartek, J. An oncogene-induced DNA damage model for cancer development. Science 319, 1352–1355 (2008).
Ten Asbroek, A. L. M. A., Van Groenigen, M., Nooij, M. & Baas, F. The involvement of human ribonucleases H1 and H2 in the variation of response of cells to antisense phosphorothioate oligonucleotides. Eur. J. Biochem. 269, 583–592 (2002).
Cerritelli, S. M. et al. Failure to produce mitochondrial DNA results in embryonic lethality in Rnaseh1 null mice. Mol. Cell 11, 807–815 (2003).
Chiba, H., Muramatsu, M., Nomoto, A. & Kato, H. Two human homologues of Saccharomyces cerevisiae SWI2/SNF2 and Drosophila brahma are transcriptional coactivators cooperating with the estrogen receptor and the retinoic acid receptor. Nucleic Acids Res. 22, 1815–1820 (1994).
de la Serna, I. L. et al. Mammalian SWI–SNF complexes contribute to activation of the hsp70 gene. Mol. Cell. Biol. 20, 2839–2851 (2000).
Méndez, J. & Stillman, B. Chromatin association of human origin recognition complex, Cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 20, 8602–8612 (2000).
García-Rubio, M., Barroso, S. I. & Aguilera, A. Detection of DNA–RNA hybrids in vivo. Methods Mol. Biol. 1672, 347–361 (2018).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Gyori, B. M., Venkatachalam, G., Thiagarajan, P. S., Hsu, D. & Clement, M. V. OpenComet: an automated tool for comet assay image analysis. Redox Biol. 2, 457–465 (2014).
Sanz, L. A. & Chédin, F. High-resolution, strand-specific R-loop mapping via S9.6-based DNA–RNA immunoprecipitation and high-throughput sequencing. Nat. Protoc. 14, 1734–1755 (2019).
Jalili, V. et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2020 update. Nucleic Acids Res. 48, 8205–8207 (2020).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Kim, D., Paggi, J. M., Park, C., Bennett, C. & Salzberg, S. L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915 (2019).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Breese, M. R. & Liu, Y. NGSUtils: a software suite for analyzing and manipulating next-generation sequencing datasets. Bioinformatics 29, 494–496 (2013).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137. (2008).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Liao, Y., Smyth, G. K. & Shi, W. FeatureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Ritchie, M. E. et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Yu, G., Wang, L. G. & He, Q. Y. ChIP seeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics https://doi.org/10.1093/bioinformatics/btv145 (2015).
Zerbino, D. R. et al. Ensembl 2018. Nucleic Acids Res. 46, D754–D761 (2018).
Ramírez, F. et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016).
Petryk, N. et al. Replication landscape of the human genome. Nat. Commun. 7, 10208 (2016).
Bianco, J. N. et al. Analysis of DNA replication profiles in budding yeast and mammalian cells using DNA combing. Methods 57, 149–157 (2012).
Robinson, J. T. et al. Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011).
Afgan, E. et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 44, W3–W10 (2016).
Gao, J. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, l1 (2013).
Gonzalez-Perez, A. et al. IntOGen-mutations identifies cancer drivers across tumor types. Nat. Methods 10, 1081–1082. (2013).
Martínez-Jiménez, F. et al. A compendium of mutational cancer driver genes. Nat. Rev. Cancer 20, 555–572 (2020).
Acknowledgements
We thank J. C. Reyes (CABIMER, Seville) for reagents provided and discussions, and the Genomic Unit of CABIMER for DNA sequencing. Research was funded by grants from the European Research Council (grant no. ERC2014 AdG669898 TARLOOP), the Spanish Ministries of Economy and Competitiveness (grant no. BFU2016-75058-P) and Science and Innovation (grant no. PDI2019-104270GB-I00/BMC), the European Union (FEDER) and Foundation ‘Vencer el Cancer’. A.B-F. was supported in part by a Juan de la Cierva postdoctoral contract (FJCI-2017-34536) from the Spanish Ministry of Science and Innovation.
Author information
Authors and Affiliations
Contributions
A.B.-F. and A.A. designed the study and the experiments. A.B.-F. performed most of the experiments and all the bioinformatic analysis. S.B. and S.M. contributed with specific experiments. A.B.-F and A.A. wrote the manuscript. All authors read, discussed and agreed with the final version of this manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Genetics thanks Stephan Hamperl and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Analysis of DNA damage and genome instability in siBRG1 cells.
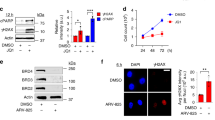
a, Quantification of nuclear BRG1 signal intensity in control (siC) and BRG1-depleted (siBRG1) HeLa cells. Data plotted as scatter plot (n = 3). Scale bar, 20 μm. (Mann-Whitney U test, two-tailed). b, WB of chromatin fraction from cells treated as in (a) using anti-BRG1 antibody (n = 2). Pounceau S membrane staining is shown as loading control. c, Quantification of depletion efficiency in cells treated as in (a) as measured by cDNA. Data are plotted as mean + SEM (n = 3). (Unpaired Student’s t-test, two-tailed). d, Representative images of γH2AX IF in control (siC) and BRG1-depleted (siBRG1) cells with (+) and without (−) RNH1 overexpression. Scale bar, 5 μm. e, Quantification of nuclear S9.6 signal intensity in control cells (siC) and BRG1-depleted cells using either a pool or individual siRNAs. Data presented as scatter plot (n = 3). Scale bar, 10 μm. (Mann-Whitney U test, two-tailed). f, Quantification of S9.6 foci per cell (left) and nucleolar signal intensity (right) in cells treated as in (d). Data plotted as scatter plot (n = 3). Representative images are shown. Scale bar, 10 μm. (Mann-Whitney U test, two-tailed). g, DRIP-qPCR expressed as % of input in cells treated as in (a). Data plotted as mean + SEM (n = 3). (Paired Student’s t-test, one-tailed). h, Quantification of nuclear S9.6 signal intensity in control cells (siC) and BRG1-depleted (siBRG1) cells transfected either with an empty plasmid (−) or a plasmid expressing an siRNA-resistant wild-type (WT) or catalytically-dead (K785R) BRG1. Data presented as scatter plot (n = 3). Scale bar, 10 μm. (Mann-Whitney U test, two-tailed). i, Percentage of cells in each cell cycle phase in cells treated as in (d). Cell cycle profiles (left) and percentages of cells for each cell cycle phase plotted as mean + SD (right) are shown (n = 3). (Paired Student’s t-test, two-tailed). P-values are indicated. Other details as in Fig. 1.
Extended Data Fig. 2 Effect of BRG1 depletion on DNA replication dynamics.
a, Quantification of nuclear EdU signal intensity in EdU+ control (siC) and BRG1-depleted (siBRG1) HeLa cells with (+) and without (−) RNH1 overexpression. Data plotted as box plot (n = 3). Scale bar, 10 μm. (Mann-Whitney U test, two-tailed). b, Representative FANCD2 IF (red) examples from cells treated as in (a) (n = 3). Scale bar, 5 μm. c, Representative BLM IF (green) examples from siC and siBRG1-treated cells (n = 2). Scale bar, 5 μm. d, Percentage of EdU-positive cells showing FANCD2 foci in siBRG1 cells. Scale bar, 10 μm. Data plotted as mean + SEM (n = 3). e, Percentage of EdU-positive cells showing BLM foci in siBRG1 cells. Scale bar, 10 μm. Data are plotted as mean + SEM (n = 3). f, Quantification of DNA (DAPI;blue) and FANCD2 (red) profiles along the indicated cell slices (yellow line) in siBRG1 cells. Scale bar, 5 μm. g, Quantification of DNA (DAPI;blue) and BLM (green) profiles along the indicated cell slices (yellow line) in siBRG1 cells. Scale bar, 5 μm. h, Representative S9.6 IF images in HeLa cells treated as in (c) along the cell cycle (n = 3). Cell phases are indicated (G1 (dark blue), S (light blue) and G2 (green)). Scale bar, 10 μm. i, Quantification of S9.6 intensity by flow cytometry in cells treated as in (c) along the cell cycle. Fold changes on S9.6 signal from G1 to S+G2/M are plotted as scatter plot (n = 3). Mean + SD are shown. (Paired Student’s t-test, one-tailed). j, Quantification of chromatin-bound BRG1 signal intensity along the cell cycle. Data plotted as box plot (n = 3). Representative images are shown. Cell cycle phases are indicated as in (h). Scale bar, 10 μm. (Mann-Whitney U test, two-tailed). k, Representative BRG1+PCNA PLA images in wild-type HeLa cells. Controls and positive PLA (red) are shown (n = 1). Scale bar, 10 μm. P-values are indicated. Other details as in Figs. 1 and 2.
Extended Data Fig. 3 Genome-wide analysis of R-loop accumulation upon BRG1 depletion.
a, DRIP-seq mean coverage over gene bodies (±2kb) from control samples (n = 2). b, DRIP-qPCR in control K562 cells. Immunoprecipitation values normalized to untreated samples. Data are mean + SEM (n = 3). (Paired Student’s t-test, one-tailed). c, DRIP-seq mean-coverage over DRIPc-seq peaks (n = 2). d, Length of siC DRIPc-seq peaks (n = 2). e, Genome annotation of DRIPc peaks of siC and R loop-gain peaks siBRG1 K562 cells. f, Vulcano plot of DRIPc-seq differential analysis between siC (blue) and siBRG1 (vermillion) K562 cells (n = 2). Dots represent peaks. g, Absolute numbers of de novo and increasing DRIPc-seq peaks of siBRG1 cells (n = 2). h, Representative screenshot comparing siC and siBRG1 DRIPc-seq profiles considering or discarding Alu-containing reads. Watson-Crick strand reads shown together (n = 2). i, Metagenomic analysis of DRIPc-seq mean coverages along gene bodies (n = 2). j, BRG1 ChIP-seq average coverage (n = 2) over genes presenting R-loop-gain peaks distributed in quartiles according to the R loop-gaining rate of DRIPc-seq. Dots represent genes. Regression line in red. k, Venn diagram with BRG1-bound and R-loop-gain genes after siBRG1 (Hypergeometric test). l, Gene lengths of entire genome and R-loop-gain genes (Mann-Whitney U-test, two-tailed). m, Expression of R-loop-gain genes in siC and BRG1-depleted cells (Mann-Whitney U-test, two-tailed). n, Box-plot of GC-content. Details as in (l). o, Metaplot of GC-skew along gene body over gene sets of (l). Dark and light-colored lines represent genes from each strand. p, Box-plot of expression values. Details as in (l). q, Metaplot of DRIPc-seq mean coverage increase (ΔDRIPc-seq) upon BRG1-depletion over chromosomes or rDNA (n = 2). r, Venn diagram showing R-loop-forming genes in siC cells and R-loop-gain genes in siBRG1 and siUAP56 K562 cells. Other details as in Figs. 1–3.
Extended Data Fig. 4 Epistatic study between BRG1 and known R loop-preventing factors.
a, Depletion controls by WB in double-knockdown experiments. siRNAs and antibodies used are indicated (n = 2). b, Representative S9.6 IF in HeLa cells transfected with the indicated siRNAs combined either with siC (−) or siBRG1 (+) (n = 3; except for siC, siTHOC1, siTHOC1+siBRG1, siUAP56 and siUAP56+siBRG1 where n = 4). Scale bar, 5 μm. c, Representative γH2AX IF in HeLa cells treated as in (b) (n = 3; except for siC and siFANCD2 where n = 4). Scale bar, 10 μm. d, Quantification of nuclear S9.6 signal intensity through cell cycle in siTHOC1 single depletion and siTHOC1+siBRG1 double depleted HeLa cells. Data presented as box plot (n = 3). Scale bar, 10 μm. (Mann-Whitney U test, two-tailed). e, Quantification of nuclear S9.6 signal intensity through cell cycle in siUAP56 single depletion and siUAP56+siBRG1 double depleted HeLa cells. Data presented as box plot (n = 3). Scale bar, 10 μm. (Mann-Whitney U test, two-tailed). f, Representative RNAPII S2P+PCNA PLA example images in wild-type HeLa cells. Control PLAs are also shown (n = 1). Scale bar, 10 μm. g, Quantification of RNAPII S2P+PCNA PLA in control cells (siC) and BRG1-depleted cells (siBRG1) either transfected with an empty plasmid (−) or a plasmid allowing expression of a siRNA-resistant wild-type (WT) or catalytically dead (K785R) version of BRG1. Data presented as scatter plot (n = 3). Scale bar, 10 μm. (Mann-Whitney U test, two-tailed). h, Representative RNAPII S2P+PCNA PLA example images in HeLa cells treated as in (b) (n = 3; except for siC (n = 6) and siSETX (n = 4)). Scale bar, 10 μm. P-values are indicated. Other details as in Figs. 1 and 2.
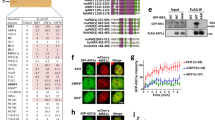
Extended Data Fig. 5 Evaluation of BRG1 occurrence at RF stalling sites.
a, Representative FANCD2+BRG1 PLA example images in wild-type HeLa cells. Control PLAs are also shown (n = 1). b, Representative FANCD2+BRG1 PLA example images in control (siC) and UAP56-depleted (siUAP56) HeLa cells with (+) and without (−) overexpression of RNH1. (n = 3). c, Representative RPA S4/8P+BRG1 PLA example images in wild-type HeLa cells. Control PLAs are also shown (n = 1). d, Representative γH2AX+BRG1 PLA example images in wild-type HeLa cells (n = 1). e, Representative RPA S4/8P+BRG1 PLA example images in siC, siUAP56 and siUAP56+siBRG1-transfected HeLa cells (n = 3). f, Representative γH2AX+BRG1 PLA example images in HeLa cells treated as in (e) (n = 3). g, Representative S9.6+BRG1 PLA example images in wild-type HeLa cells (n = 1). h, Representative S9.6+BRG1 PLA example images in HeLa cells treated as in (b) (n = 3). Scale bar, 10 μm. Other details as in Fig. 1.
Extended Data Fig. 6 BRG1 genome-wide co-localization analysis with R loop, RF stalling and DNA damage sites.
a, Venn diagram showing BRG1 (yellow), FANCD2 (red) and γH2AX (blue) target genes co-occurrence in control K562 cells. b, DRIPc-seq (green), BRG1 (yellow), FANCD2 (red) and γH2AX (purple) ChIP-seq data mean coverage around T-R conflicts (±1Mb). Data presented as metaplot according to RF direction. c, Quantification of co-directional (CD; green bar) vs head-on (HO; red bar) T-R conflicts. Absolute numbers are plotted. d, BRG1 (yellow) and FANCD2 (red) ChIP-seq data mean coverage around HO T-R conflicts (±1Mb). Data presented as metaplot according to RF direction. e, As in (d), but around CD T-R conflicts (±1Mb). f, DRIPc-seq average coverage at DRIPc-seq peaks reached by the RF in CD or HO orientation during DNA replication. Data plotted as box plot. (Mann-Whitney U test, two-tailed). g, BRG1 ChIP-seq average coverage at genes reached by the RF in HO and CD orientation. Data plotted as box plot. (Mann-Whitney U test, two-tailed). h, FANCD2 ChIP-seq average coverage at genes reached by the RF in HO and CD orientation. Data plotted as box plot. (Mann-Whitney U test, two-tailed). P-values are indicated. Other details as in Figs. 2 and 6. Average coverages from two DRIPc-seq and BRG1 ChIP-seq biological replicates are shown.
Extended Data Fig. 7 Chromatin accessibility and nucleosome occupancy analysis at R loop gain sites in BRG1-deficient cells.
a, Correlation analysis between K562 and HAP1 BRG1 ChIP-seq analysis. b, Correlation analysis between K562 and CD36 BRG1 ChIP-seq analysis. c, Correlation analysis between BRG1 ChIP-seq in K562 and BIN67 (BRG1-deficient) cells expressing BRG1. d, Representative γH2AX IF example images in HeLa cells transfected with the indicated siRNAs and either en empty plasmid (−) or a plasmid for RNH1 overexpression (+). Scale bar, 10 μm. e, Representative S9.6 IF example images in HeLa cells transfected with the indicated siRNAs and treated as in (d). Scale bar, 10 μm. f, Representative screenshot comparing siC (blue) and siBRG1 (vermillion) DRIPc-seq data in K562 and ChIP-seq profiles of BRG1 (yellow), BRM (green), ARID1A (orange) and PBRM1 (purple) in HAP1 cells. g, BRG1, BRM, ARID1A and PBRM1 ChIP-seq mean coverage in HAP1 WT cells over genes identified as R loop-gain genes in siBRG1 K562 (± 2kb). Data plotted as metagene. h, BRG1, BRM, ARID1A and PBRM1 ChIP-seq mean coverage in HAP1 WT cells around R loop accumulating peaks in siBRG1 K562 cells (± 10kb). i, HAP1 WT, BRG1 KO, BRM KO, ARID1A KO and PBRM1 KO ATAC-seq average coverage at R loop gain sites identified in siBRG1 K562 cells. Data plotted as box plot. (Mann-Whitney U test, two-tailed). j, HAP1 WT, BRG1 KO, BRM KO, ARID1A KO and PBRM1 KO ATAC-seq mean coverage around HO (left) and CD (right) T-R collisions (± 1Mb). Data plotted as heatmap according to RF direction. Pearson correlations (r) are shown in (a), (b), and (c). P-values are indicated in (i). Other details as in Figs. 2 and 6.
Extended Data Fig. 8 SWI/SNF mutation and cancer.
a, Number of identified mutations in cancer from a curated set on non-redundant cancer studies (www.cbioportal.com) for the main SWI/SNF (red), ISWI (yellow), CHD (blue) and INO80 (green) ATPase subunits. b, Number of cancer types where the gene is considered a cancer driver gene (www.intogen.org) for the main SWI/SNF (red), ISWI (yellow), CHD (blue) and INO80 (green) ATPase subunits. c, Distribution of cancer-associated BRG1 mutations along the protein. Protein length (grey) and domains (colored squares) are drawn. Highly frequently and recurrently altered in cancer protein residues (cancer hotspots) are highlighted as orange dots in the upper track. K785R point mutation used in rescue experiments is indicated. d, Quantification of nuclear S9.6 signal intensity in HeLa (BRG1-proficient) and C-33 A (BRG1-deficient) cells. Data presented as scatter plot (n = 3). Scale bar, 10 μm. (Mann-Whitney U test, two-tailed). e, Number of identified mutations in cancer from a curated set on non-redundant cancer studies (www.cbioportal.com) for the main SWI/SNF genes. f, Number of cancer types where the gene is considered a cancer driver gene (www.intogen.org) for the main SWI/SNF genes. P-values are indicated. Other details as in Fig. 1.
Supplementary information
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 7
Statistical source data.
Source Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 1
Unprocessed western blots.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 4
Unprocessed western blots.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 8
Statistical source data.
Rights and permissions
About this article
Cite this article
Bayona-Feliu, A., Barroso, S., Muñoz, S. et al. The SWI/SNF chromatin remodeling complex helps resolve R-loop-mediated transcription–replication conflicts. Nat Genet 53, 1050–1063 (2021). https://doi.org/10.1038/s41588-021-00867-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-021-00867-2
This article is cited by
-
Unscheduled excessive R-loops in immune response
Functional & Integrative Genomics (2024)
-
In vivo dynamics and regulation of DNA G-quadruplex structures in mammals
Cell & Bioscience (2023)
-
SWI/SNF complexes in hematological malignancies: biological implications and therapeutic opportunities
Molecular Cancer (2023)
-
The chromatin-associated RNAs in gene regulation and cancer
Molecular Cancer (2023)
-
Genome-wide measurement of DNA replication fork directionality and quantification of DNA replication initiation and termination with Okazaki fragment sequencing
Nature Protocols (2023)