Abstract
The ongoing COVID-19 pandemic has caused a global economic and health crisis. To identify host factors essential for coronavirus infection, we performed genome-wide functional genetic screens with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and human coronavirus 229E. These screens uncovered virus-specific as well as shared host factors, including TMEM41B and PI3K type 3. We discovered that SARS-CoV-2 requires the lysosomal protein TMEM106B to infect human cell lines and primary lung cells. TMEM106B overexpression enhanced SARS-CoV-2 infection as well as pseudovirus infection, suggesting a role in viral entry. Furthermore, single-cell RNA-sequencing of airway cells from patients with COVID-19 demonstrated that TMEM106B expression correlates with SARS-CoV-2 infection. The present study uncovered a collection of coronavirus host factors that may be exploited to develop drugs against SARS-CoV-2 infection or future zoonotic coronavirus outbreaks.
Similar content being viewed by others
Main
The COVID-19 pandemic, caused by SARS-CoV-2, has resulted in a worldwide health crisis1 and few effective drugs are available to treat patients with COVID-19. Although remdesivir initially seemed promising for severe cases2, the World Health Organization’s Solidarity trial showed that it has no definite impact on mortality3. Dexamethasone can reduce mortality by a third among critically ill patients with COVID-19, by suppressing the hyperactive immune response4. However, as treatment benefits severe cases only to a limited extent, efficient and safe therapeutics are urgently required while awaiting the worldwide implementation of vaccines.
Coronaviruses cause respiratory and intestinal infections in a broad range of mammals and birds. Seven human coronaviruses (HCoVs) are known, which probably all emerged as zoonoses from bats, mice or domestic animals5. The four so-called ‘common cold HCoVs’—229E, NL63, OC43 and HKU1—cause mild upper respiratory tract illnesses6. In contrast, SARS-CoV, Middle East respiratory syndrome coronavirus (MERS-CoV) and the recently emerged SARS-CoV-2 are highly pathogenic and cause severe, potentially lethal respiratory infections. As numerous coronaviruses reside in animal reservoirs and interspecies transmission frequently occurs5,7,8, there is a constant risk of new pathogenic coronaviruses spreading into the human population, as exemplified by the recent SARS-CoV-2 pandemic. Nevertheless, our options to prevent or treat coronavirus infections remain limited. Hence, the development of broad-spectrum anti-coronavirus drugs could help not only to address the current high medical need, but also to quickly contain zoonotic events in the future. Common host factors essential for replication of multiple coronaviruses represent attractive targets for broad-spectrum antiviral drugs.
To develop such drugs, it is crucial to understand which host factors coronaviruses require to infect a cell, because each step of the coronavirus replication cycle (receptor binding, endocytosis, fusion, viral protein translation, genome replication, virion assembly and release) may serve as a target for intervention. Although the entry step of coronaviruses has been relatively well characterized, the host–virus interplay in later steps of the viral life cycle remains largely elusive. For SARS-CoV-2, previous studies have shown that the protein angiotensin-converting enzyme 2 (ACE2) can serve as a receptor in Vero E6 cells9 or in human cells overexpressing ACE2 (refs. 10,11,12). In addition, it was shown that the SARS-CoV-2 spike (S) can be primed for fusion by cellular proteases such as furin, transmembrane serine protease 2 (TMPRSS2) or cathepsin B or L, depending on the target cell type10,13.
In the present study, we performed a series of genome-wide CRISPR (clustered regularly interspaced short palindromic repeats)-based genetic screens to identify host factors required for SARS-CoV-2 and HCoV-229E infection. We identified phosphoinositide 3-kinase (PI3K) type 3 as a common host factor for SARS-CoV-2, HCoV-229E and HCoV-OC43, and show that small molecules targeting this protein might serve as broadly applicable anti-coronavirus inhibitors. Furthermore, we discovered that the lysosomal protein TMEM106B serves as an essential specific host factor for SARS-CoV-2 infection in multiple liver- and lung-derived human cell lines.
Results
Genome-wide knockout screens for HCoV-229E and SARS-CoV-2
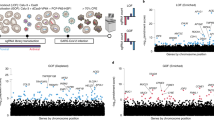
Genome-wide knockout screens have been widely used to identify host factors for various viruses14,15 and were recently performed with coronaviruses, including SARS-CoV-2 (refs. 9,12,16,17,18,19). Most of these screens were performed in human cells that were engineered to overexpress ACE2 (refs. 12,16,18,19). In the present study, we performed a CRISPR-based genome-wide knockout screen in the human Huh7 cell line, without introducing an exogenous receptor, because our SARS-CoV-2 strain induced a clear cytopathic effect (CPE) in these cells. We performed screens with both SARS-CoV-2 and the less pathogenic HCoV-229E. This allowed for the identification of both (1) broad-spectrum coronavirus host factors and (2) specific host factors for SARS-CoV-2 and HCoV-229E. Huh7 cells transduced with the Brunello genome-wide library20 were selected for survival during infection with either coronavirus. Single guide (sg)RNAs present in the resistant cell population were identified by deep sequencing (Fig. 1a). We performed high-stringency screens for HCoV-229E (Fig. 1b) and SARS-CoV-2 (Fig. 1c) by exposing cells to the virus until nearly all the cells had died. For SARS-CoV-2, we also performed a lower-stringency screen (Fig. 1d) to identify genes having a more subtle effect on virus infection.
a, Overview of experimental steps performed during a genome-wide screen for coronavirus host factors. b–d, Genome-wide knockout screens performed in Huh7 cells, with strong selection (high stringency) using HCoV-229E (b) and SARS-CoV-2 (c) or with mild SARS-CoV-2 selection (low stringency) (d). Each circle represents a gene, with size corresponding to significance (−log(P value)) of enrichment. The enrichment (y axis) represents the average change in abundance of the four sgRNAs for each gene, when comparing sgRNA abundances between the virus-selected population and the sgRNA library before selection (b and c) or between the virus-selected population and an uninfected control population (d). Genes are distributed on the x axis in alphabetical order. The sgRNA-level P values were calculated using a one-sided Student’s t-test and aggregated using Fisher’s method to obtain gene-level P values44. Data result from two independent screen replicates.
As expected, the HCoV-229E screen identified ANPEP (Fig. 1b), encoding the established HCoV-229E receptor aminopeptidase N (AP-N)21. Our SARS-CoV-2 screens did not identify ACE2, in contrast to screens performed in Vero E6 cells9 or ACE2-overexpressing human cells12,16,18. This is probably due to the low ACE2 expression in Huh7 cells compared with Vero E6 or ACE2-overexpressing cells22 (Extended Data Fig. 1). The high-stringency SARS-CoV-2 screen (Fig. 1c) identified one significantly enriched gene, TMEM106B, encoding the poorly characterized protein TMEM106B, which is involved in lysosome function and implicated in neurodegenerative disorders23. A larger number of genes were enriched in the low-stringency screen (Fig. 1d). However, a low-stringency screen may have an increased background due to the presence of a subpopulation of cells that are not infected and may be selected for resistance to antiviral stress signals rather than resistance to virus. Two genes were present among the top-ranked genes in both the HCoV-229E and the SARS-CoV-2 low-stringency screen (Fig. 1b,d): PIK3C3 (endocytic trafficking and autophagy) and TMEM41B (autophagy). Among the top 40 most enriched genes in the low-stringency SARS-CoV-2 screen, there were two genes required for cell surface receptor expression (EXT1 and ITGB6), three lipid homeostasis genes (OSBPL9, PTDSS1 and TMEM30A) and three additional genes involved in membrane/vesicle trafficking (CCZ1B, PTPN23 and RSG1). As viruses frequently employ factors involved in membrane biology, we focused on the 11 genes described above for further validation.
Genetic screen validation
To validate the findings from our genetic screens, we expressed sgRNAs targeting the identified genes and the known receptor genes ANPEP and ACE2 in Huh7 cells, and tested whether their ablation affected the sensitivity of cells to CPE induced by HCoV-229E or SARS-CoV-2. On-target activity of sgRNAs was checked by sequencing (Extended Data Fig. 2). As expected, sgRNAs targeting ANPEP protected cells from HCoV-229E-induced cell death (Fig. 2a). Cells were only partially protected from SARS-CoV-2-induced CPE by sgRNAs targeting ACE2, which is in line with the absence of ACE2 in our screens. The only highly enriched gene in our high-stringency SARS-CoV-2 screen was TMEM106B. The sgRNAs targeting TMEM106B protected Huh7 cells from CPE caused by SARS-CoV-2, even when exposed to high virus concentrations, but had no effect on cells infected with HCoV-229E (Fig. 2a).
a, Huh7 cells expressing control sgRNAs (targeting safe harbor AAVS1), gene-specific pools of four sgRNAs or individual sgRNAs infected with SARS-CoV-2 dilutions (sixfold) or HCoV-229E dilutions (tenfold), incubated 3 d at 35 °C and stained with crystal violet. One of two repetitions is shown. b, Huh7 cells expressing control sgRNAs (targeting safe harbor AAVS1), gene-specific pools of four sgRNAs, or the two best performing individual sgRNAs from the screens, infected with SARS-CoV-2 (MOI = 0.2). Cell viability was determined by MTS assay 2 d post-infection (control (AAVS1), n = 24; OSBPL9 pool, CCZ1B sgRNA 4, n = 4; PTDSS1 pool, PTPN23 pool, RSG1 pool, n = 3; for all other conditions n = 6, from two independent experiments). Error bars denote mean ± s.e.m. Data were analyzed using Brown–Forsythe and Welch ANOVA (two sided) and Dunnett’s T3 multiple comparison test, comparing each experimental condition with the control. Significant increases in viability compared with control (AAVS1) P values are indicated on the graph.
We further tested sgRNAs targeting the ten selected genes from the low-stringency screen, as well as TMEM106B, using a quantitative readout (Fig. 2b). Besides TMEM106B, the knockout of which conferred the strongest resistance to SARS-CoV-2, significant resistance was observed for two individually tested sgRNAs targeting PIK3C3, TMEM41B, EXT1, OSBPL9 or TMEM30A. For CCZ1B and RSG1, resistance was observed only when using a pool of four sgRNAs. In both wild-type Huh7 cells (Extended Data Fig. 3a) and cells expressing ACE2 sgRNAs (Extended Data Fig. 3b), knockout of EXT1, but not ITGB6, protected cells against SARS-CoV-2-induced CPE. EXT1 encodes exostosin-1 which is involved in the synthesis of heparan sulfate; this places this finding in line with a recent report showing that SARS-CoV-2 infection can be mediated by heparan sulfate22. OSBPL9 encodes oxysterol-binding protein-related protein 9 (ORP-9), which mediates transport of sterols between the endoplasmic reticulum and the trans-Golgi network24. TMEM30A encodes transmembrane protein 30A, a subunit of the ATP8A2 flippase complex that regulates the distribution of phospholipids across membrane leaflets25. Together, these results demonstrate that SARS-CoV-2 infection requires TMEM106B as well as genes involved in heparan sulfate synthesis (EXT1), autophagy (PIK3C3 and TMEM41B) and lipid transport (OSBPL9 and TMEM30A).
PI3K type 3 is a druggable common anti-coronavirus target
PIK3C3 encodes PI3K type 3, the catalytic subunit of the PI3K complex that mediates the formation of phosphatidylinositol 3-phosphate and plays a role in many processes, including endocytic trafficking and the initiation and maturation of autophagosomes26. As inhibitors directly targeting this protein are available, we tested the activity of the structurally distinct inhibitors VPS34-IN1, VPS34-IN2, SAR405 and autophinib against different coronaviruses (Fig. 3a–d). As expected, all PI3K type 3 inhibitors inhibited the formation of LC3-positive autophagosome puncta and induced large vacuoles in treated cells (Extended Data Fig. 4a)27. PI3K type 3 inhibitors showed antiviral activity against SARS-CoV-2 (Fig. 3a,c) and were also active against HCoV-229E and HCoV-OC43 (Fig. 3b,d). As PI3K type 3 is involved in autophagosome formation, we investigated whether macroautophagy is required for SARS-CoV-2 and HCoV-229E infection. Disruption of ATG5 and ATG7, required for phagophore expansion28, did not affect the induction of CPE by SARS-CoV-2 or HCoV-229E (Fig. 3e), whereas cells were unable to form LC3-positive autophagosomes (Extended Data Fig. 4b), confirming disruption of the macroautophagy pathway. These results show that SARS-CoV-2 and other coronaviruses employ PI3K type 3 for infection but do not depend on a functional macroautophagy pathway.
a, eGFP-expressing Vero E6 cells pretreated for 24 h with indicated compounds and infected with SARS-CoV-2. Cell viability without virus was determined by MTS assay 4 d after compound treatment. Cell viability with virus was calculated by measuring the number of eGFP-positive cells compared with uninfected, untreated controls (n = 2 wells from one of two independent experiments with similar results). b, Huh7 cells pretreated for 30 min with indicated compounds and infected with HCoV-229E. Cell viability with/without virus was determined by MTS assay 3 d post-infection and/or compound treatment (viability data with virus: n = 3 independent experiments for VPS34-IN1; n = 4 for VPS34-IN2, SAR405 and autophinhib; viability data without virus: n = 3 independent experiments for VPS34-IN1; n = 4 for autophinhib; n = 5 for VPS34-IN2 and SAR405). c,d, Huh7 cells infected with SARS-CoV-2 (c) or HRT-18G cells infected with HCoV-OC43 (d). Relative number of infected cells compared with untreated control after treatment with PI3K type 3 inhibitors for 6 h, as determined by high-content image analysis after dsRNA staining (n = 6 wells examined over two independent experiments). Cell viability without virus was determined by MTS assay 6 h post-infection (n = 4 wells examined over two independent experiments). e, Huh7 cells expressing pools of four sgRNAs targeting ATG5, ATG7 or safe harbor AAVS1 infected with dilutions of SARS-CoV-2 or HCoV-229E. Cell viability was determined by MTS assay after 3 d (n = 6 wells examined over two independent experiments). Area under the curve (AUC) values were calculated and analyzed using Brown–Forsythe and Welch ANOVA and Dunnett’s T3 multiple comparison test, comparing each experimental condition with the control. No significant differences were found. f, Vero E6 cells preincubated for 30 min with a concentration series of the indicated compounds and subsequently infected with pseudoparticles harboring the SARS-CoV-2 spike (either wild-type or deletion; see also Extended Data Fig. 4d) (n = 4 wells examined over two independent experiments). In parallel, cell viability without pseudovirus was determined by MTS assay 1 d after compound treatment. Data are expressed as percentage GFP-positive cells or percentage viability over the mean of untreated controls. wt, wild-type. Fit curves were calculated by least squares regression (a, b, e and f). Error bars denote mean ± s.e.m. CC50 and IC50 ± s.e.m. are half-maximal cytotoxic (CC50) and inhibitory (IC50) concentrations.
A recent study reported that deletions near the proteolytic cleavage site at the S1/S2 boundary in the SARS-CoV-2 spike protein can affect the mode of cell entry12. As such a deletion is present in our SARS-CoV-2 strain (SARS-CoV-2/Belgium/GHB-03021/2020), we tested activity of PI3K type 3 inhibitors against entry of pseudoparticles harboring wild-type SARS-CoV-2 spike or SARS-CoV-2 spike containing deletions in the S1 domain and a deletion near the S1/S2 cleavage site (Extended Data Fig. 4d). PI3K type 3 inhibitors blocked infection with both pseudoviruses (Fig. 3f), showing that this dependency is not affected by the aforementioned spike deletions. A time-of-drug-addition experiment showed that inhibition of HCoV-229E by SAR405 occurs later in the viral life cycle than the attachment stage, as benchmarked by the attachment inhibitor Urtica dioica agglutinin (UDA), but earlier than onset of intracellular replication as identified using the viral RNA synthesis inhibitor remdesivir (Extended Data Fig. 4c). The above data suggest a role for PI3K type 3 in an early step of the viral life cycle, but downstream of receptor binding.
TMEM41B is a crucial host factor for HCoV-229E infection
TMEM41B, which is involved in the early stage of autophagosome formation29, was identified as a host factor in both our HCoV-229E and our SARS-CoV-2 screens (Fig. 1b,d). Although TMEM41B knockout almost completely protected Huh7 cells against CPE induced by HCoV-229E (Fig. 4a,b), the effect on SARS-CoV-2-induced CPE was subtle (Figs. 2b and 4a). The protective effect of TMEM41B knockout was reverted by complementation with TMEM41B complementary DNA, confirming specificity (Fig. 4a). In line with this effect on cell viability, release of infectious HCoV-229E virions was reduced in TMEM41B knockout cells, whereas no significant effect on release of SARS-CoV-2 virions was observed (Fig. 4c). TMEM41B knockout or overexpression did not affect infection with pseudoparticles harboring the HCoV-229E spike protein (Fig. 4d), suggesting a role of TMEM41B in a post-entry step. A recent study also suggested a post-entry role of TMEM41B in flavivirus replication complex formation30. Together, these data identify the autophagy protein TMEM41B as an indispensable host factor for HCoV-229E.
a, Huh7 cells expressing control sgRNAs (AAVS1), a TMEM41B sgRNA pool (TMEM41BKO) and/or TMEM41B cDNA infected with SARS-CoV-2 or HCoV-229E. Cell viability was determined by MTS assay after 3 d at 35 °C (n = 6 wells examined over two independent experiments; n = 3 TMEM41BKO, MOI = 0.003). b, Huh7 cells expressing a TMEM41B sgRNA pool or separate sgRNAs infected with HCoV-229E. Cell viability was determined by MTS assay after 3 d at 35 °C (n = 3 wells from one independent experiment; fit curves were calculated by least squares regression). a,b, AUC values were calculated and analyzed using Brown–Forsythe and Welch ANOVA and Dunnett’s multiple comparison test, comparing each experimental condition with the control. P values are indicated for significant differences (two sided) relative to the control (colored as in graph). c, Huh7 cells expressing a TMEM41B sgRNA pool (TMEM41BKO) infected with SARS-CoV-2 or HCoV-229E. The amount of infectious virus in the supernatant was determined at different times post-infection by end-point dilution data (n = 3 wells from one independent experiment) and was log(transformed) and analyzed by two-way ANOVA with Geiser–Greenhouse correction and Dunnett’s multiple comparison test, comparing each experimental condition with the control. Values significantly different from AAVS1 control are marked as follows: **0.001 <P <0.01; *0.01 < P <0.05. d, Huh7 cells expressing a TMEM41B sgRNA pool (TMEM41BKO) or TMEM41B cDNA infected with pseudoparticles harboring the HCoV-229E spike (n = 6 wells examined over two independent experiments). Data are expressed as percentage GFP-positive cells, normalized to wild-type cells. Error bars denote mean ± s.e.m. Data were analyzed using Brown–Forsythe and Welch ANOVA and Dunnett’s multiple comparison test, comparing each experimental condition with the control. No significant differences were found.
TMEM106B is required for SARS-CoV-2 infection
Our high-stringency SARS-CoV-2 screen identified an important role for TMEM106B. We further investigated the effect of TMEM106B knockout on SARS-CoV-2 infection in multiple human cell lines. TMEM106B knockout in Huh7 and Hep3B cells prevented SARS-CoV-2-induced CPE (Fig. 5a and Extended Data Fig. 5a) and reduced virus replication, as measured by staining of double-stranded (ds) RNA replication intermediates (Fig. 5b and Extended Data Fig. 5b). Complementation with sgRNA-resistant TMEM106B cDNA restored CPE and infectivity (Fig. 5a,b and Extended Data Fig. 5a–c), confirming specificity of TMEM106B knockout. Knockout and complementation of TMEM106B were verified by immunostaining (Extended Data Fig. 5d,e). It is interesting that overexpression of TMEM106B in Huh7 cells accelerated the onset of CPE and enhanced SARS-CoV-2 infection (Fig. 5a,b). Such enhancement of infection was also observed for another SARS-CoV-2 strain BavPat-1 (ref. 31), which lacks deletions in the spike protein (Extended Data Fig. 4d), as well as for the related SARS-CoV, the causative agent of SARS outbreaks in 2003 (Extended Data Fig. 6a).
a, Huh7 cells expressing control sgRNAs (AAVS1), a pool of four sgRNAs targeting TMEM106B and/or TMEM106B cDNA infected with sixfold dilutions of SARS-CoV-2 and stained with crystal violet after 3 d at 35 °C. b, Huh7 cells expressing a pool of four sgRNAs targeting TMEM106B and/or TMEM106B cDNA infected with SARS-CoV-2 strain BetaCov/Belgium/GHB-03021/2020 and stained for dsRNA after 6 h (n = 6 wells examined over two independent experiments). Data are expressed as percentage infected cells, normalized to control cells (AAVS1) and analyzed using Brown–Forsythe and Welch ANOVA with Dunnett’s T3 multiple comparison, comparing each experimental condition with the control; P values are shown for significant differences (two sided). c, Lung-derived cell lines (A549 and NCI-H1975) or primary HBECs expressing control sgRNAs (AAVS1) or a pool of four sgRNAs targeting TMEM106B infected with sixfold dilutions of SARS-CoV-2 and stained with crystal violet after 8 d (A549) at 35 °C or 6 d (NCI-H1975) or 3 d (HBECs) at 37 °C. d, NCI-H1975 wild-type or monoclonal TMEM106B knockout cells transduced with TMEM106B cDNA, infected with sixfold dilutions of SARS-CoV-2 and stained with crystal violet after 3 d at 35 °C. e, NCI-H1975 wild-type or monoclonal TMEM106B knockout cells transduced with TMEM106B cDNA, infected with SARS-CoV-2 and stained for dsRNA after 6 h (n = 6 wells examined over two independent experiments). Data are expressed as percentage infected cells, normalized to control cells (AAVS1). ND, not detectable. Data were analyzed using Brown–Forsythe and Welch ANOVA with Dunnett’s T3 multiple comparison, comparing each experimental condition with the control. P values are shown for significant differences (two sided). f, NCI-H1975 wild-type or monoclonal TMEM106B knockout cells infected with SARS-CoV-2 or HCoV-229E, and infectious virus in supernatant quantified by end-point dilution (n = 3 wells from one experiment). g, NCI-H1975 wild-type or monoclonal TMEM106B knockout cells infected with SARS-CoV-2 at MOI = 1. Cells were stained with crystal violet and viral RNA in supernatant measured by qPCR at the indicated time points (n = 3 wells from one experiment). f,g, Data were log transformed and analyzed by two-way ANOVA with Geiser–Greenhouse correction and Bonferroni’s or Dunnett’s multiple comparison test, comparing each experimental condition with the control. Values significantly different (two sided) from the control are marked (****P < 0.0001; ***0.0001 < P < 0.001; **0.001 < P < 0.01). h, Huh7 cells expressing a TMEM106B sgRNA pool (TMEM106BKO) or TMEM106B cDNA transduced with the indicated pseudoparticles harboring the SARS-CoV-2 (either wild-type or deletion), SARS-CoV or HCoV-229E spike (n = 6 wells examined over two independent experiments). WT, wild-type. Data are expressed as percentage GFP-positive cells, normalized to control cells (AAVS1). Error bars denote mean ± s.e.m. Data are analyzed using Brown–Forsythe and Welch ANOVA with Dunnett’s T3 multiple comparison, comparing each experimental condition with the control. P values are shown for significant differences (two sided).
In addition to liver-derived cell lines Huh7 and Hep3B, we expressed TMEM106B sgRNAs in lung-derived cell lines NCI-H2110, A549, NCI-H1975 and primary human bronchial epithelial cells (HBECs). TMEM106B knockout cells were consistently resistant to SARS-CoV-2-induced CPE (Fig. 5c and Extended Data Fig. 6b). To further corroborate the importance of TMEM106B for SARS-CoV-2 infection, we generated monoclonal NCI-H1975 cells from a polyclonal TMEM106B knockout population. Complete knockout of TMEM106B protected cells from SARS-CoV-2-induced CPE (Fig. 5d) and prevented infection (Fig. 5e), which were both restored by complementation with TMEM106B cDNA. Furthermore, no increase in replication-competent virus progeny (Fig. 5f) or viral RNA (Fig. 5g and Extended Data Fig. 6e) was observed in the supernatant of infected TMEM106B knockout cells, whereas HCoV-229E production was not affected in these cells (Fig. 5f). These data further underscore the essential nature of TMEM106B for SARS-CoV-2 infection.
To investigate whether TMEM106B is required for SARS-CoV-2 spike-mediated entry, Huh7 cells either expressing TMEM106B sgRNAs or overexpressing TMEM106B were infected with pseudoparticles containing SARS-CoV-2 spike. Although infection of control Huh7 was inefficient and no significant effect of TMEM106B knockout was detected, TMEM106B overexpression stimulated SARS-CoV-2 pseudovirus infection (Fig. 5h). This effect was observed for both wild-type spike proteins and spike containing the aforementioned deletions (Extended Data Fig. 4d), but also for SARS-CoV spike protein. In contrast, it was not observed for spike from TMEM106B-independent HCoV-229E. Altogether, these results demonstrate that TMEM106B is a bona fide proviral host factor required for productive SARS-CoV-2 infection of human cells and suggest a role of TMEM106B in the entry stage of the replication cycle.
TMEM106B expression correlates with SARS-CoV-2 infection
To further investigate the relevance of TMEM106B for SARS-CoV-2 infection, we analyzed TMEM106B expression in a single-cell RNA-sequencing (scRNA-seq) dataset of bronchoalveolar lavages from 44 patients with pneumonia32; 31 patients (84,874 cells) were confirmed COVID-19 positive by reverse transcription quantitative PCR (RT–qPCR) and 13 (31,923 cells) were negative and served as the control. In this dataset, we further identified cells infected by SARS-CoV-2 using the Viral Track method, which detects SARS-CoV-2-derived sequencing reads in scRNA-seq data33. In these 116,797 cells, TMEM106B expression was detected in 4,094 (18%) epithelial cells (versus 8% and 13% of lymphoid and myeloid cells, respectively; Fig. 6a). It is interesting that epithelial cells from patients with COVID-19 showed a significantly increased TMEM106B expression compared with epithelial cells from controls (Fig. 6b). This increase was especially evident in SARS-CoV-2-infected cells, when assessing both sequencing reads of the S viral protein (indicative of viral replication) and all viral sequences (Fig. 6b). Subclustering epithelial cells revealed that this increase was significant for ciliated, hillock and secretory epithelial cells (Fig. 6c). These data demonstrate that SARS-CoV-2-susceptible airway cells of patients with COVID-19 express increased levels of TMEM106B.
a, UMAP representation of 116,797 cells obtained from bronchoalveolar lavage of non-COVID-19 (n = 13) and COVID-19 (n = 31) patients, analyzed by scRNA-seq. Cells are color coded for the indicated cell type (left) and color coded for log(normalized TMEM106B expression) (right). b,c, Violin plots showing TMEM106B expression per cell subtype (b) and epithelial cell subtype (c) stratified for control (non-COVID-19), COVID-19 (all cells from patients with COVID-19), SARS-CoV-2 (cells from patients with COVID-19 containing any viral sequence detected) or SARS-CoV-2 spike (cells from patients with COVID-19 containing sequence encoding S detected), with partial overlap between stratified populations. To obtain log normalized expression levels (a, b and c), the read count corresponding to TMEM106B for each cell was divided by the total read count for that cell and multiplied by 10,000. Values were then natural log transformed (log(1P function)). A Kruskal–Wallis test (P values shown in black) was performed to test whether differences occur within each cell type (b) or subtype (c). For groups with significant differences, pairwise comparisons were performed using Wilcoxon’s rank-sum test (P values shown in red).
Discussion
Although several coronavirus receptors and proteolytic activators of the S protein have been extensively studied, little remains known about coronavirus host factor requirements. In the present study, we performed genome-wide genetic screens with the human coronaviruses HCoV-229E and SARS-CoV-2. These screens identified virus-specific as well as common host factors, such as PI3K type 3, which we validated as a druggable host factor shared among SARS-CoV-2, HCoV-229E and HCoV-OC43. We show that the autophagy regulator TMEM41B is required for HCoV-229E infection, and to a lesser extent for SARS-CoV-2 infection. Finally, we identified TMEM106B as a crucial cellular host factor for SARS-CoV-2 infection and showed that TMEM106B expression correlates with SARS-CoV-2 infection in airway epithelial cells.
Our high-stringency screen identified TMEM106B as the strongest dependency factor for SARS-CoV-2 infection, whereas the low-stringency screen detected multiple weaker dependencies. From these genes, we selected a subset involved in receptor biology, autophagy, membrane/vesicle trafficking and lipid homeostasis for validation. Although we could validate most of these genes, a few were not validated. The presence of such false positives might be explained by the lower stringency, resulting in the presence of uninfected cells that persisted in the population. These cells may have been selected for resistance to bystander effects such as antiviral stress signals, rather than virus infection, which does not occur in the uninfected reference population.
Simultaneous with our screens, six other groups have performed similar genome-wide CRISPR screens to identify host factors for SARS-CoV-2 (refs. 9,12,16,17,18,19) or common cold coronavirus replication16,17. The top-ranked genes from our HCoV-229E screen (ANPEP, PIK3C3 and TMEM41B) were also identified in the other screens16,17. SARS-CoV-2 screens were performed in three different cell types: the African green monkey Vero E6 cell line9, the alveolar basal epithelial carcinoma A549 cell line overexpressing ACE2 (A549ACE2)12,18,19 or the hepatoma cell line Huh7 (the present study), or derived clones Huh7.5 (ref. 17) and Huh7.5.1 overexpressing ACE2 and TMPRSS2 (Huh7.5.1ACE2/TMPRSS2)16. We compared the overlap between the 50 top-ranked genes identified in each screen, revealing 33 genes that were identified in at least 2 screens (Supplementary Tables 1 and 2). Of these, 8 were discovered in our screen, 11 in the Huh7.5 and Huh7.5.1ACE2/TMPRSS2 screens (Wang et al.16 and Schneider et al.17), 18 (Daniloski et al.18 and Zhu et al.12) or 1 (Heaton et al.19) in the A549ACE2 screens, and 5 in the Vero E6 screen (Wei et al.9). Only ACE2 was identified in four screens, whereas CTSL, EXT1, ARPC4 and TMEM106B were found in three screens, the latter three also being found in our screen. The remaining 28 genes were identified in only 2 different screens. This limited overlap between screens may be due to variations in technical setup (CRISPR library, multiplicity of infection (MOI), virus strain or cell line). In particular, screens performed in similar cell types demonstrated strong overlap (Extended Data Fig. 7).
Three genes found in our SARS-CoV-2 screens (TMEM106B, EXT1 and TMEM41B) were also found in other screens in Huh7.5(.1) cells16,17, independently confirming our results. Although our top hit TMEM106B was not identified in the A549 screens, we confirmed the effect of TMEM106B knockout on SARS-CoV-2 in this cell type (Fig. 5c). A possible explanation for the absence of TMEM106B in these screens could be that TMEM106B might not be required when cells are saturated with high ACE2 levels. Although we observed an effect of TMEM106B knockout in A549ACE2+ cells, ACE2 levels may have been lower in our cells (Extended Data Fig. 6d) than A549ACE2+ cells used in the screens12,18. TMEM106B was not identified by the screen in Vero E6 cells9, possibly because these are not of human origin. Three top-ranked genes from our screens (PIK3C3, ARPC4 and CCZ1B) overlap with top-ranked genes in at least one of the two screens in A549ACE2+ cells12,18, and two genes (OSBPL8 and PHIP) overlap with the Vero E6 screen by Wei et al.9. PIK3C3 and CCZ1B were also identified in screens with HCoV-229E and HCoV-OC43 (ref. 16). Besides the genes described above, we identified and validated OSBPL9 and TMEM30A as two additional SARS-CoV-2 host factors that were not identified by any other screen (Fig. 2b). Altogether, this shows that understanding SARS-CoV-2 host factor dependencies requires combined data from multiple genetic screens with varying experimental approaches.
All human cell lines tested in our study were susceptible to SARS-CoV-2 infection, despite very low ACE2 expression levels, suggesting possible ACE2-independent entry routes. We found that the heparan sulfate synthesis gene EXT1 is required for infection (Extended Data Fig. 3), in line with a recent study showing that heparan sulfate facilitates SARS-CoV-2 binding to cells22. Another recent study showed that receptor proteins KREMEN1 and ASGR1 can facilitate infection of SARS-CoV-2 spike pseudoparticles34. Therefore, it is plausible that SARS-CoV-2, similarly to other viruses, has a broader repertoire of cellular receptors than initially postulated.
Genes identified in both our HCoV-229E and our SARS-CoV-2 screens were PIK3C3 and TMEM41B. We validated TMEM41B as a host factor for both viruses, but found that HCoV-229E depends more strongly on this factor. Infection of TMEM41B knockout cells with HCoV-229E spike pseudovirus suggested a role for TMEM41B in a post-entry step (Fig. 4d). We further validated PIK3C3 genetically (Fig. 2b) and showed activity of PI3K type 3 inhibitors against SARS-CoV-2, HCoV-229E and HCoV-OC43 (Fig. 3a–d), demonstrating the potential of PI3K type 3 to serve as a pan-coronavirus drug target. Our data (Fig. 4f and Extended Data Fig. 4c) suggest that PI3K type 3 facilitates an early step of the viral life cycle, such as endocytosis, fusion, translation or replication initiation. Others reported that PI3K type 3 inhibitors cause dispersal of the SARS-CoV-2 N protein and dsRNA throughout the cytoplasm, suggesting a role of this factor in replication complex formation35. We showed that disrupting autophagy genes ATG5 and ATG7, required for phagophore expansion, does not block SARS-CoV-2 and HCoV-229E infection (Fig. 3e). Thus, PI3K type 3 might support infection by inducing phagophore nucleation, whereas later stages of macroautophagy are unnecessary.
Of the host factors identified in the present study, TMEM106B was the most essential for SARS-CoV-2 infection. TMEM106B is a 274 amino acid transmembrane protein that resides in endosomes and lysosomes, controlling lysosome size, number, mobility and trafficking. TMEM106B is poorly characterized and only recently received attention because of its role in frontotemporal dementia, the second leading cause of pre-senile neurodegeneration23. We showed that TMEM106B overexpression specifically enhances cell entry by pseudoviruses carrying SARS-CoV-2 or SARS-CoV spike (Fig. 5h), whereas entry mediated by spike protein from HCoV-229E, which is TMEM106B independent (Fig. 5f), was unaffected. TMEM106B plays a pivotal role in lysosomal acidification, by interacting with the proton pump vacuolar, ATPase accessory protein 1 (AP1)36. Therefore, a role of TMEM106B might be to promote endosomal acidification, to facilitate delivery of the SARS-CoV-2 genome into the cytoplasm. This is suggested by reports showing that entry of SARS-CoV-2 S-pseudotyped virus requires endosomal acidification10,37. It is interesting that HCoV-229E also requires endosomal acidification38, but does not require TMEM106B for infection (Figs. 2a and 5f), suggesting that different, although related, viruses may depend on distinct factors to exploit similar cellular pathways. Alternatively, TMEM106B may be an endosomal receptor for SARS-CoV-2, similar to the lysosomal receptor NPC1 used by Ebola virus39. Recently, multiple SARS-CoV-2 variants with increased transmissibility have emerged. Variant VOC-202012/01 from the United Kingdom, which has rapidly spread to numerous countries, contains at least 24 different mutations40, which are absent in the strains used in the present study (except spike deletion HV69-70). Future research should establish whether these mutations affect the requirement of TMEM106B and other host factors.
The relevance of TMEM106B for SARS-CoV-2 infection is evident from our scRNA-seq analysis of airway cells, which shows higher TMEM106B expression levels in airway epithelium from patients with COVID-19 compared with noninfected patients (Fig. 6). This may suggest that TMEM106B expression increases the susceptibility to SARS-CoV-2, causing higher infection rates in individuals with high TMEM106B levels. This would be in line with our in vitro data showing increased infectivity on TMEM106B overexpression (Fig. 5b,h). It would be interesting to study whether TMEM106B could serve as prognostic biomarker for severity of COVID-19. Alternatively, SARS-CoV-2 infection could stimulate TMEM106B expression in infected cells/tissues. TMEM106B expression in lung tissue is lower than in other tissues, such as the brain or testes41. Patients with COVID-19 sometimes have neurological symptoms such as stroke, brain hemorrhage and memory loss42. Furthermore, a recent study demonstrated impaired sperm quality in patients with COVID-19 (ref. 43). Therefore, it may be worth investigating whether TMEM106B plays a role in SARS-CoV-2 infection of these tissues.
As new pathogenic coronaviruses periodically emerge, these viruses will continue to pose a public health threat beyond the ongoing COVID-19 pandemic. The set of essential coronavirus host factors identified in our genome-wide screens will form a basis for studies unraveling the cellular pathways hijacked by these viruses. In addition, these factors can serve as targets for medicinal chemistry efforts to counter the current COVID-19 pandemic or future outbreaks of pathogenic coronaviruses.
Methods
Chemicals and reagents
Reference inhibitor compounds autophinib and VPS34-IN1 were purchased from Selleckchem and SAR405 and VPS34-IN2 were obtained from MedChemExpress. Plant lectin UDA isolated from the Urtica dioica rhizomes was kindly donated by E. Van Damme (Ghent, Belgium). Chloroquine was purchased from Acros Organics and remdesivir was ordered from MedKoo. Stock solutions were prepared in dimethyl sulfoxide (DMSO).
Cell culture
HEK293T (American Type Culture Collection (ATCC), catalog no. CRL-3216), Vero E6 (ATCC, catalog no. CRL-1586), Huh7 (CLS-300156; human hepatoblastoma), Hep3B (ATCC, catalog no. HB-8064; human hepatocellular carcinoma), HRT-18G (ATCC, catalog no. CRL-11663; human colorectal adenocarcinoma) and BHK-21J (received from P. Bredenbeek, LUMC, The Netherlands) were maintained in Dulbecco’s modified Eagle’s medium (DMEM, Gibco Life Technologies) supplemented with 8% heat-inactivated fetal bovine serum (FBS; HyClone, GE Healthcare Life Sciences), 0.075% sodium bicarbonate (Gibco Life Technologies) and 1 mM sodium pyruvate (Gibco Life Technologies). A549 (ATCC, catalog no. CCL-185) cells were maintained in F-12K medium supplemented with 10% heat-inactivated FBS. NCI-H1975 (ATCC, catalog no. CRL-5908) and NCI-H2110 (ATCC, catalog no. CRL-5924) cells were maintained in RPMI medium supplemented with 10% heat-inactivated FBS. All cell lines were maintained at 37 °C under 5% CO2. Primary HBECs were collected as previously described45. Ethical approval was given by the Ethical Committee of University Hospital UZ Leuven (S63978) and informed consent by the subjects/patients. Briefly, primary HBECs were isolated by overnight pronase digestion of the main-stem bronchus from a donor lung before transplantation. After digestion, primary HBECs were seeded in T75 flasks with Pneumacult Ex-Plus medium (StemCell Technologies) supplemented with 100 U ml−1 of penicillin–streptomycin and 2.5 µg ml−1 of amphotericin B until confluence (±6 d). The primary HBECs were trypsinized with 0.5% trypsin–ethylenediaminetetraacetic acid (Gibco) in RPMI-1640 medium (Gibco) and either seeded immediately in a six-well plate for gene editing or cryopreserved in 10% DMSO (Sigma-Aldrich)/FBS (Gibco) until needed. The monoclonal NCI-H1975 TMEM106B knockout cell line was generated from the polyclonal cell line that was transduced with a pool of the four sgRNAs targeting TMEM106B. First, the guides present in this cell line were identified by sequencing, and next the genomic locus targeted by these guides was sequenced. This monoclonal cell line contains two of these guides (sgRNA 1 and sgRNA 3).
Generation of virus stocks
All infectious SARS-CoV-2-related work was conducted in the high-containment BSL3+ facilities of the KU Leuven Rega Institute (3CAPS) under licenses AMV 30112018 SBB 219 2018 0892 and AMV 23102017 SBB 219 2017 0589, according to institutional guidelines. SARS-CoV-2 strain SARS-CoV-2/Belgium/GHB-03021/2020 (GenBank accession no. MW368439—passage 5) was used for all experiments, unless otherwise indicated. This strain was recovered from a nasopharyngeal swab taken from an RT–qPCR-confirmed asymptomatic patient returning from Wuhan in February 2020 (ref. 46). Infectious virus was isolated and multiplied by five serial passages on Huh7 cells. Cells were seeded in DMEM supplemented with 10% heat-inactivated FBS to reach a confluency of ~80% the next day. After replacing the medium by DMEM + 2% or 4% FBS, cells were infected with SARS-CoV-2 at an MOI of ~0.01. When most cells were dying, supernatant was removed from the cells, centrifuged to remove cell debris and stored at −80 °C. SARS-CoV-2 strain SARS-CoV-2/Germany/BavPat1/2020 (ref. 31) (GenBank accession no. MW368440—passage 2) was obtained from C. Drosten, Charité Universitätsmedizin Berlin and passaged once on Vero E6 cells. The SARS-CoV Frankfurt 1 strain was obtained from H. F. Rabenau of the Johann Wolfgang Goethe University, Frankfurt, Germany and was propagated in Vero E6 cells. The HCoV-229E (ATCC, catalog no. VR-740) and HCoV-OC43 (ATCC, catalog no. VR-1558) virus stocks were obtained by inoculating a confluent monolayer of Huh7 or HRT-18G cells, respectively. The supernatant was harvested after 3 d of incubation for HCoV-229E, or 7 d of incubation for HCoV-OC43, at 35 °C under 5% CO2, and stored in aliquots at −80 °C after one freeze–thaw cycle and removal of cellular debris by centrifugation.
Genome-wide knockout screens
For the HCoV-229E and SARS-CoV-2 (high-stringency) screens, 1.5 × 108 Huh7 cells for each of two replicates were transduced at an MOI of ~0.3 with lentivirus containing the Brunello genome-wide library in lentiCRISPRv2 (Addgene, catalog no. 73179), which contains 77,441 sgRNAs targeting 19,114 genes. Cells were selected with 2 µg ml−1 of puromycin for 3 d to eliminate untransduced cells, seeded at a coverage of ~200 cells per sgRNA for each replicate and infected with HCoV-229E (MOI = 0.5) or SARS-CoV-2 (MOI = 2.5). Surviving cells were harvested at 18 d post-infection (HCoV-229E) or 41 d post-infection (SARS-CoV-2). For the SARS-CoV-2 low-stringency screen, 1.5 × 108 Huh7 cells for each of two replicates were transduced at an MOI of ~0.2 with lentivirus containing the Brunello library and selected with puromycin for 3 d. Then, cells were seeded at a coverage of 500 cells per sgRNA for each replicate in DMEM with 4% FBS and infected with SARS-CoV-2 at an MOI of 0.1. For each replicate, uninfected cells were maintained under similar conditions to the infected cells and harvested simultaneously. Then, 5 d post-infection, cells were cultivated in DMEM with 20% FBS for 3 d to allow cell recovery, and were infected again with an MOI of ~0.1. Cells were harvested at 14 d post-infection. Genomic DNA was extracted from cells using the QIAmp DNA Mini Kit (QIAGEN, catalog no. 51306) or, for the SARS-CoV-2 low-stringency screen, with the QIAmp DNA Blood maxi kit (QIAGEN, catalog no. 51194). In a first PCR step, regions of ~600 bp containing the sgRNA sequence were amplified using NEBNext Ultra II Q5 Master Mix (New England Biolabs, catalog no. M0544S) in 25 amplification cycles. A second PCR of 10 cycles with NEBNext Ultra II Q5 Master Mix was performed with primers containing Illumina adapters and TruSeq indices. Products were separated by agarose gel electrophoresis and purified with the PureLink Quick Gel Extraction Kit (Thermo Fisher Scientific, catalog no. K210012). Samples were then diluted to 2–4 nM, pooled, and denatured and diluted according to the instructions for single-end sequencing on an MiSeq (Illumina) with an MiSeq-v.2 50 cycles or an MiSeq-v.3-150 cycles kit (Illumina) and 10% PhiX (Illumina) spike-in. FastQ files were further analyzed with CRISPRCloud2 (ref. 44), using the Brunello library sgRNA list (Supplementary Table 3). In this analysis, the gene-level enrichment is obtained by averaging the sgRNA-level enrichment values. The sgRNA-level P values are calculated using a one-sided Student’s t-test and aggregated using Fisher’s method to obtain gene-level P values. Bubble plots of screen data were generated using Microsoft Excel 2016. Read counts and gene-level analyses of all screens are listed in Supplementary Tables 4–11.
Genome-wide knockout screen hit validation
For individual validation of genes, guides enriched during the genome-wide knockout screens were cloned into the pLentiCRISPRv2 plasmid (Addgene, catalog no. 52961) following the standard cloning protocol. For lentiviral particle production, HEK293T cells were plated in 40 ml of supplemented DMEM in T150 (TPP) flasks at 45% confluency and incubated overnight. After 24 h, the cells were transfected using X-TremeGENE 9 (Roche) with the pLentiCRISPR plasmids and the lentiviral packaging plasmids pMD2.G and psPAX2 to generate lentiviral particles coated with the vesicular stomatitis virus G (VSV-G) protein and incubated overnight. Then, 24 h post-transfection the medium was changed to DMEM supplemented with serum-free bovine serum albumin growth medium (DMEM + 1.1 g 100 ml−1 of bovine serum albumin and 20 µg l−1 of gentamicin). The supernatant containing lentiviral particles was harvested 72 h after transfection and stored at −80 °C. Cells were transduced with lentiviruses expressing only one sgRNA or a pool of the four sgRNAs from the Brunello genome-wide knockout library and then selected with puromycin for 3 d (sgRNAs target sequences are in Supplementary Table 12). Monoclonal TMEM106B knockout cells were derived from polyclonal cells expressing an sgRNA pool, by seeding a dilution series of cells and selecting wells containing a single cell colony. Knockout was confirmed by Sanger sequencing of sgRNA target sites and staining with a TMEM106B-specific antibody.
DNA extraction and sequencing
Genomic DNA was isolated from 1 million cells with the QIAamp DNA Mini Kit using RNase A. For Sanger sequencing, the region of interest was amplified using the CloneAmp HiFi PCR premix (Clontech) in 25-µl PCR reactions containing 50 ng of genomic DNA. The amplified DNA was then purified (Nucleospin Gel and PCR Clean-up (Macherey-Nagel)) and sequenced (Macrogen; primer sequences are in Supplementary Table 12) and analyzed using Geneious software (v.9.1.8).
Cell viability assays
Cells stably expressing specific sgRNAs were seeded in 96-well plates at 4,000 cells per well in medium with 8% or 10% FBS. The following day, serial dilutions of virus in medium without FBS were added to the cells, resulting in a serum concentration of 4% or 5%. Cells were incubated until sufficient CPE was visible. For MTS assays, medium was removed from the cells and replaced by MTS reagent (CellTiter 96 AQueous One Solution Cell Proliferation Assay from Promega) diluted in phosphate-buffered saline (PBS). The absorbance was measured with a Tecan Spark microplate reader. For crystal violet staining, cells were fixed in 4% formaldehyde for 30 min, stained with a 1% crystal violet solution in water and rinsed with water.
Virus production assays
Cells were infected with virus for 1 h at 35 °C in DMEM without FBS. After infection, the inoculate was replaced by DMEM with 4% FBS. Supernatant was harvested at different time points. The amount of infectious virus was determined by end-point dilution on Vero E6 cells (for SARS-CoV-2) or Huh7 cells (for HCoV-229E). The values of the 50% tissue culture infectious doses (TCID50) per ml were calculated using the Spearman–Kärber method. The amount of viral RNA in supernatant was quantified by qPCR as follows: RNA was extracted from the supernatant, reverse-transcribed and PCR amplified using the QIAprep& Viral RNA UM Kit (QIAGEN, catalog no. 221415) according to the manufacturer’s instructions. SARS-CoV-2 replication was assessed using the SARS-CoV-2 N1+N2 Assay Kit (QIAGEN, catalog no. 222015). Amplification and detection were performed in an QuantStudio 5 Real-Time PCR System (Applied Biosystems). Ct (cycle threshold) values were converted to relative RNA levels and normalized to the RNA level in samples harvested immediately after infection.
Virus inhibition assays
The antiviral activity of PI3K type 3 inhibitors on SARS-CoV-2 in Vero E6 cells was evaluated as follows: on day −1, the test compounds were serially diluted in DMEM (Gibco, catalog no. 41965-039) supplemented with 2% (v:v) heat-inactivated fetal calf serum and sodium bicarbonate (Gibco, catalog no. 25080-060). Diluted compounds were then mixed with enhanced green fluorescent protein (eGFP)-expressing Vero E6 cells at 25,000 cells per well in 96-well plates (Greiner Bio-One, catalog no. 655090). The plates were incubated overnight in a humidified incubator at 37 °C and 5% CO2. On day 0, SARS-CoV-2 was added at 20 TCID50 per well and, on day 4 post-infection, the wells were examined for eGFP expression using a high-content imaging platform, and the images of the wells were converted into signal values. To obtain values for cell viability (with virus), the background signal was subtracted based on infected, untreated controls, and signal values were normalized to uninfected, untreated controls. The toxicity of compounds in the absence of virus was evaluated by MTS assay. All compounds were tested in duplicate, in two independent experiments. To evaluate the antiviral activity of PI3K type 3 inhibitors against HCoV-229E, Huh7 cells were seeded into 384-well plates. The next day, serial dilutions of the compounds were added to the cells before infection with HCoV-229E at 30 TCID50 per well. At 3 d post-infection, the virus-induced CPE was measured by MTS assay.
Time-of-drug-addition assay
Huh7 cells were seeded into 48-well dishes at 40,000 cells per well. After 24 h of incubation at 37 °C, the cells were cooled on ice for 1 h, followed by addition of 30 CCID50 of the HCoV-229E virus and further incubation at 35 °C. The test compounds were added at a concentration approximately twofold above their half-maximal effective concentration (EC50), at different time points post-infection (p.i.): −30 min, 0 h, 30 min, 1 h, 2 h, 3 h, 5 h and 8 h p.i. At 11 h p.i., total cellular RNA extracts were prepared and viral RNA was quantified using the CellsDirect One-Step qRT-PCR kit (Thermo Fisher Scientific). One-step real-time RT-PCR was performed using the 229E-FP forward primer, 229E-RP reverse primer and TaqMan minor groove-binder probe 229E-TP (Thermo Fisher Scientific; Supplementary Table 12), derived from the HCoV-229E membrane protein gene sequence as described previously47. Amplification and detection were performed in an ABI 7500 Fast Sequence Detection System (Applied Biosystems) under the following conditions: an initial reverse transcription at 50 °C for 15 min, followed by PCR activation at 95 °C for 2 min and 45 cycles of amplification (15 s at 95 °C and 30 s at 60 °C). Six independent experiments were carried out.
Immunofluorescence assays
Immunofluorescence staining was performed according to standard procedures. Briefly, all cells were seeded at a density of 20,000 cells per well in 8-well µ-slides (Ibidi). Cells were allowed to adhere overnight before receiving compound treatment and/or viral infection with SARS-CoV-2 or HCoV-OC43. After incubation, cells were fixed (4% paraformaldehyde in PBS), washed and permeabilized (0.2% Triton X-100 in PBS). Employed primary antibodies were rabbit anti-LC3B (Sigma-Aldrich, catalog no. L7543) at a 1:200 dilution, mouse anti-dsRNA (J2, Scicons) at a 1:1,000 dilution and rabbit anti-TMEM106B (Novus, catalog no. NBP2-31714) at a 1:50 dilution. Secondary antibodies Alexa Fluor-568 goat anti-rabbit (A11011, Invitrogen, Thermo Fisher Scientific) and Alexa Fluor-488 goat anti-mouse (A11029, Invitrogen, Thermo Fisher Scientific) were diluted 1:500. Cell nuclei were counterstained with DAPI and the samples were imaged by confocal microscopy on a Leica TCS SP5 confocal microscope (Leica Microsystems), employing an HCX PL APO 63× (numerical aperture 1.2) water immersion objective. The percentage of infected cells was quantified by high-content image analysis (ArrayScan XTI, Thermo Fisher Scientific) for at least 3,000 cells per condition.
Generation of cells overexpressing ACE2, TMEM106B and TMEM41B
The pLCKO plasmid was a gift from J. Moffat (Addgene, plasmid no. 73311). The invariant gRNA scaffold was removed together with the puromycin resistance gene and replaced by the coding sequence of ACE2 (Addgene, plasmid no. 1786), TMEM106B (Integrated DNA Technologies) or TMEM41B (Integrated DNA Technologies), followed by a P2A-coupled blasticidin resistance gene driven by a cytomegalovirus promoter. Protospacer adjacent motif sequences flanking binding sites of sgRNAs from the Brunello library and the sgRNA target sequences were mutagenized in the coding sequence of TMEM106B and TMEM41B by introducing silent mutations. The resulting vector was used to make lentiviral particles, as described above. Huh7 cells were transduced with the lentiviral stock in the presence of polybrene (8 µg ml−1). After 24 h, medium was replaced by medium containing blasticidin (10 µg ml−1) and cells were incubated for an additional 48 h.
Simple Western analysis
For Simple Western analysis, cells were lysed in RIPA lysis buffer (Sigma-Aldrich) for 1 h at 4 °C. Whole cell lysates were cleared by centrifugation. Proteins were separated by size (12–230 kDa) and visualized on a Wes system (ProteinSimple) with an anti-mouse or anti-goat immunoglobulin G–horseradish peroxidase antibody (R&D Systems, catalog no. HAF109, diluted 1:5) detecting the primary antibody against glyceraldehyde 3-phosphate dehydrogenase (Santa Cruz Biotechnology, catalog no. sc-47724, diluted 1:500), vinculin (Cell Signaling, catalog no. 13901, diluted 1:500) or anti-hACE2 (R&D Systems, catalog no. AF933, diluted 1:400). Protein signals were visualized and quantified with the Compass software, v.4.0.0 (ProteinSimple).
Pseudotyping of VSV
Expression plasmids encoding SARS-CoV-S and SARS-CoV-2-S were obtained from B. J. Bosch and W. Li, Utrecht University, The Netherlands. Plasmids encoding HCoV-229E-S and MERS-S were from S. Pöhlmann, DPZ, Göttingen, Germany. Mutations in the S sequence were introduced via PCR and inserted into pCAGGS using the NEBuilder HiFi DNA Assembly kit. VSV pseudotypes were generated as described previously10. Briefly, HEK293T or BHK-21J cells were transfected with the respective expression plasmids; 1 d p.i., cells were infected (MOI = 2) with VSVΔG expressing a GFP reporter gene (purchased from Kerafast) for 2 h. The medium was changed with medium containing anti-VSV-G antibody (I1-hybridoma, ATCC, catalog no. CRL-2700) to neutralize residual VSV-G input. After 24 h, supernatant containing VSV pseudotypes was harvested. For transduction, the indicated cell lines were inoculated with the respective pseudotyped VSV. For experiments involving inhibitors, cells were preincubated for 30 min at 37 °C with serial dilutions of the indicated compounds. After 18 h of incubation, the percentage of GFP-expressing cells was quantified on a Cell Insight CX5/7 High Content Screening platform (Thermo Fisher Scientific) with Thermo Fisher Scientific HCS Studio (v.6.6.0) software.
Flow cytometry
Cell surface expression of ACE2 protein was detected by flow cytometry. Briefly, confluent cells were trypsinized and left to rest at 37 °C for 90 min. Cells were resuspended in PBS with 2% FBS (assay buffer) containing 1 μg ml−1 of anti-hACE2 antibody (R&D Systems, catalog no. AF933, 1:10 dilution) and incubated for 30 min. Cells were washed twice, resuspended in assay buffer containing 0.5 μg ml−1 of PE-conjugated donkey anti-goat (LifeSpan BioSciences, catalog no. LS-C752241, 1:200 dilution) and incubated for another 30 min. Cells were washed twice and resuspended in PBS with 1% paraformaldehyde for FACS analysis using a BD FACSCanto II Flow Cytometer.
Analysis of scRNA-seq data
We assessed the TMEM106B expression levels in an integrated single-cell dataset consisting of 116,797 cells32,48. These cells were stratified among control, COVID-19- and SARS-CoV-2-infected cells from patients with COVID-19 (COVID-19 infected). The COVID-19-infected cells were identified by Viral Track33. Differences among the control, COVID-19 and COVID-19-infected cells were statistically assessed using a Kruskal–Wallis test, and P < 0.05 was considered significant. For groups with significant differences, pairwise comparisons were performed using Wilcoxon’s rank-sum test. We visualized these cells using a Uniform Manifold Approximation and Projection (UMAP) and violin plots using the Seurat49 package v.3 (https://github.com/satijalab/seurat/releases/tag/v3.0.0) in R version 3.6.0—‘Planting of a Tree’. The code used to generate Fig. 6 is available at https://github.com/u0054600/COVID19-TMEM106B.
Statistical analysis
Statistical analyses of all data, except genetic screen and single-cell sequencing data, were performed using GraphPad Prism software. Data were analyzed for normality and log(normality) using the Kolmogorov–Smironov test before application of analysis of variance (ANOVA). All analyses were performed at a threshold α level of 0.05. The specific statistical tests used for each dataset are mentioned in the respective figure captions, as well as any data transformation that was applied. All measurements were taken from distinct samples.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Sequencing data are available from the National Center for Biotechnology Information Sequencing Read Archive (SRA accession nos. SRR13255539, SRR13255540, SRR13255541, SRR13255542, SRR13255543, SRR13255544, SRR13255545, SRR13255546, SRR13255547; BioProject accession no. PRJNA685335). The scRNA-seq data are from another study32; raw sequencing reads have been deposited in the EGA European Genome–Phenome Archive database (EGAS00001004717) and a download of the processed read count matrix is available at http://covid19.lambrechtslab.org.
Code availability
The code used to generate Fig. 6 is available at https://github.com/u0054600/COVID19-TMEM106B.
References
Dong, E., Du, H. & Gardner, L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 20, 533–534 (2020).
Beigel, J. H. et al. Remdesivir for the treatment of Covid-19—preliminary report. N. Engl. J. Med. https://doi.org/10.1056/NEJMoa2007764 (2020).
WHO Solidarity Trial Consortium et al. Repurposed antiviral drugs for Covid-19—interim WHO solidarity trial results. N. Engl. J. Med. https://doi.org/10.1056/NEJMoa2023184 (2020).
Horby, P. et al. Effect of dexamethasone in hospitalized patients with COVID-19: preliminary report. N. Engl. J. Med. https://doi.org/10.1056/NEJMoa2021436 (2020).
Ye, Z. W. et al. Zoonotic origins of human coronaviruses. Int. J. Biol. Sci. 16, 1686–1697 (2020).
Liu, D. X., Liang, J. Q. & Fung, T. S. Human Coronavirus-229E, -OC43, -NL63, and -HKU1. Ref. Modul. Life Sci. https://doi.org/10.1016/b978-0-12-809633-8.21501-x (2020).
Chan, J. F., To, K. K., Tse, H., Jin, D. Y. & Yuen, K. Y. Interspecies transmission and emergence of novel viruses: lessons from bats and birds. Trends Microbiol. 21, 544–555 (2013).
Cui, J., Li, F. & Shi, Z. L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 17, 181–192 (2019).
Wei, J. et al. Genome-wide CRISPR screens reveal host factors critical for SARS-CoV-2 infection. Cell https://doi.org/10.1016/j.cell.2020.10.028 (2020).
Hoffmann, M. et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell https://doi.org/10.1016/j.cell.2020.02.052 (2020).
Zhou, P. et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 (2020).
Zhu, Y. et al. The S1/S2 boundary of SARS-CoV-2 spike protein modulates cell entry pathways and transmission. Preprint at bioRxiv https://doi.org/10.1101/2020.08.25.266775 (2020).
Shang, J. et al. Cell entry mechanisms of SARS-CoV-2. Proc. Natl Acad. Sci. USA 117, 11727–11734 (2020).
Flint, M. et al. A genome-wide CRISPR screen identifies N-acetylglucosamine-1-phosphate transferase as a potential antiviral target for Ebola virus. Nat. Commun. 10, 1–13 (2019).
Li, B. et al. Genome-wide CRISPR screen identifies host dependency factors for influenza A virus infection. Nat. Commun. 11, 164 (2020).
Wang, R. et al. Genetic screens identify host factors for SARS-CoV-2 and common cold coronaviruses. Cell https://doi.org/10.1016/j.cell.2020.12.004 (2020).
Schneider, W. M. et al. Genome-scale identification of SARS-CoV-2 and pan-coronavirus host factor networks. Cell https://doi.org/10.1016/j.cell.2020.12.006 (2020).
Daniloski, Z. et al. Identification of required host factors for SARS-CoV-2 infection in human cells. Cell https://doi.org/10.1016/j.cell.2020.10.030 (2020).
Heaton, B. E. et al. SRSF protein kinases 1 and 2 are essential host factors for human coronaviruses including SARS-CoV-2. Preprint at bioRxiv https://doi.org/10.1101/2020.08.14.251207 (2020).
Doench, J. G. et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR–Cas9. Nat. Biotechnol. 34, 184–191 (2016).
Yeager, C. L. et al. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature 357, 420–422 (1992).
Clausen, T. M. et al. SARS-CoV-2 infection depends on cellular heparan sulfate and ACE2. Cell 183, P1043–P1057 (2020).
Nicholson, A. M. & Rademakers, R. What we know about TMEM106B in neurodegeneration. Acta Neuropathol. 132, 639–651 (2016).
Ngo, M. & Ridgway, N. D. Oxysterol binding protein-related protein 9 (ORP9) is a cholesterol transfer protein that regulates Golgi structure and function. Mol. Biol. Cell 20, 1388–1399 (2009).
Coleman, J. A. & Molday, R. S. Critical role of the beta-subunit CDC50A in the stable expression, assembly, subcellular localization, and lipid transport activity of the P4-ATPase ATP8A2. J. Biol. Chem. 286, 17205–17216 (2011).
Backer, J. M. The intricate regulation and complex functions of the class III phosphoinositide 3-kinase Vps34. Biochem. J. 473, 2251–2271 (2016).
Ronan, B. et al. A highly potent and selective Vps34 inhibitor alters vesicle trafficking and autophagy. Nat. Chem. Biol. 10, 1013–1019 (2014).
Dikic, I. & Elazar, Z. Mechanism and medical implications of mammalian autophagy. Nat. Rev. Mol. Cell Biol. 19, 349–364 (2018).
Morita, K. et al. Genome-wide CRISPR screen identifies TMEM41B as a gene required for autophagosome formation. J. Cell Biol. 217, 3817–3828 (2018).
Hoffmann, H. et al. TMEM41B is a pan-flavivirus host factor. Cell https://doi.org/10.1016/j.cell.2020.12.005 (2020).
Wölfel, R. et al. Virological assessment of hospitalized patients with COVID-2019. Nature 581, 465–469 (2020).
Wauters, E. et al. Discriminating mild from critical COVID-19 by innate and adaptive immune single-cell profiling of bronchoalveolar lavages. Cell Res. https://doi.org/10.1101/2020.07.09.196519 (2021).
Bost, P. et al. Host-viral infection maps reveal signatures of severe COVID-19 patients. Cell 181, 1475–1488.e12 (2020).
Gu, Y. et al. Interaction network of SARS-CoV-2 with host receptome through spike protein. Preprint at bioRxiv https://doi.org/10.1101/2020.09.09.287508 (2020).
Silvas, J. A., Jureka, A. S., Nicolini, A. M., Chvatal, S. A. & Basler, C. F. Inhibitors of VPS34 and lipid metabolism suppress SARS-CoV-2 replication. Preprint at bioRxiv https://doi.org/10.1101/2020.07.18.210211 (2020).
Klein, Z. A. et al. Loss of TMEM106B ameliorates lysosomal and frontotemporal dementia-related phenotypes in article loss of TMEM106B ameliorates lysosomal and frontotemporal dementia-related phenotypes in progranulin-deficient mice. Neuron 95, 281–296.e6 (2017).
Ou, X. et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 11, 1620 (2020).
Blau, D. M. & Holmes, K. V. Human coronavirus HCoV-229E enters susceptible cells via the endocytic pathway. Adv. Exp. Med. Biol. 494, 193–198 (2001).
Carette, J. E. et al. Ebola virus entry requires the cholesterol transporter Niemann–Pick C1. Nature 477, 340–343 (2011).
Leung, K. et al. Early empirical assessment of the N501Y mutant strains of SARS-CoV-2 in the United Kingdom, October to November 2020. Euro. Surveill. 26, 2002106 (2021).
Uhlén, M. et al. Tissue-based map of the human proteome. Science 347, 1260419 (2015).
Varatharaj, A. et al. Neurological and neuropsychiatric complications of COVID-19 in 153 patients: a UK-wide surveillance study. Lancet Psychiatry 7, 875–882 (2020).
Li, H. et al. Impaired spermatogenesis in COVID-19 patients. EClinicalMedicine 28, 100604 (2020).
Jeong, H. H., Kim, S. Y., Rousseaux, M. W. C., Zoghbi, H. Y. & Liu, Z. Beta-binomial modeling of CRISPR pooled screen data identifies target genes with greater sensitivity and fewer false negatives. Genome Res. 29, 999–1008 (2019).
Mathyssen, C. et al. Vitamin D modulates the response of bronchial epithelial cells exposed to cigarette smoke extract. Nutrients 11, 2138 (2019).
Boudewijns, R. et al. STAT2 signaling restricts viral dissemination but drives severe pneumonia in SARS-CoV-2 infected hamsters. Nat. Commun. 11, 5838 (2020).
Vijgen, L. et al. Development of one-step, real-time, quantitative reverse transcriptase PCR assays for absolute quantitation of human coronaviruses OC43 and 229E. J. Clin. Microbiol. 43, 5452–5456 (2005).
Liao, M. et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 26, 842–844 (2020).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019).
Acknowledgements
We thank N. Van Winkel and L. Mercelis for help with infection experiments, N. Willems and J. Punjwani for help with transductions, immunofluorescence staining and cell culture, K. Minner for help with cloning and stable cell line generation, L. Bral for help with qPCR, cloning and stable cell line generation, B. Massant for help with immunofluorescence staining, cloning, sequencing and stable cell line generation, and N. Thys for pseudotyped virus experiments. We thank the European Virus Archive for providing the BavPat strain. S.J., J.N. and K.D. were supported by the Research Foundation Flanders under the Excellence of Science program (VirEOS project no. 30981113). J.B. is supported by a KU Leuven PDM grant (no. PDM/19/168). J.W. is supported by an FWO Fundamental Clinical Mandate (no. 1833317N). E.W. is supported by Stichting tegen Kanker (mandate for basic and clinical oncology research). The scRNA-seq data were funded by the Grand Challenges Program of VIB from the Flemish Government under the Management Agreement 2017-2021 (VR 2016 2312 Doc.1521/4) to D.L. This project has received funding from the FWO Hercules Foundation (grant no. ZW13-02), Caps-It infrastructure.
Author information
Authors and Affiliations
Contributions
J.B., S.J. and P.M. performed genetic screens. J.B., L.P., S.J. and J.D.M. performed infectivity assays. L.P., E.V. and M.J. performed other experiments. D.V.L. and H.J.T performed assays involving pseudoviruses. J.W. and E.W. provided airway epithelial cells for scRNA-seq. B.B. analyzed scRNA-seq data. V.G. prepared primary HBECs. J.B., L.P., E.V., M.J., D.V.L. and H.J.T. were involved in data analysis. J.B., L.P., E.V., M.J. and D.D designed the project. L.P, D.J., J.N., K.D., B.M.V., D.L., P.M. and D.D. supervised and supported the project. J.B., E.V., M.J. and D.D. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Genetics thanks Alexander Ploss and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Analysis of ACE2 expression levels in different cell lines.
Lysates of the indicated wildtype cell lines, or Huh7 cells transduced with an ACE2 overexpression construct, were analyzed using a ProteinSimple Wes™ system, with antibodies specific for ACE2 and the endogenous controls vinculin and GAPDH. One of two repetitions is shown.
Extended Data Fig. 2 Genotyping of CRISPR/Cas9-mediated gene knockout for validation of host factors for SARS-CoV-2 and HCoV-229E.
Forty-eight hours after transduction of cells with lentivirus containing sgRNAs, target loci of sgRNAs were PCR-amplified, Sanger sequenced and aligned to the wildtype (WT) reference sequence. The sgRNA sequence is highlighted in red, the Cas9 PAM sequence is highlighted in blue and the cleavage site is indicated by a red arrowhead. The monoclonal NCI-H1975 TMEM106B knock out cell line was generated from the polyclonal cell line that was transduced with a pool of 4 sgRNAs targeting TMEM106B. sgRNAs present in this monoclonal cell line were first identified by sequencing and subsequently the genomic locus targeted by these guides was sequenced. This monoclonal cell line contains two guides (sgRNA 1 and sgRNA 3). Mismatches in the sequencing chromatogram are highlighted in blue.
Extended Data Fig. 3 SARS-CoV-2 infection requires the heparan sulfate biosynthesis factor EXT1.
a,b, Huh7 wildtype cells (a) or Huh7 cells expressing a pool of sgRNAs targeting ACE2 (b) were transduced with lentivirus containing indicated sgRNA pools and subsequently infected with a dilution series of SARS-CoV-2 and incubated for three days at 35 °C, followed by measurement of cell viability by MTS assay. Data are mean ± s.e.m. Fit curves were calculated by least squares regression. AUC values were calculated and analysed by Brown-Forsythe and Welch ANOVA and Dunnet’s multiple comparison test, comparing each experimental condition to the control. For significant differences from the control (two-sided), p values are indicated in the figure (colored as in graph legend) (n = 3 wells from one experiment).
Extended Data Fig. 4 Coronavirus infection requires PI3K type 3 in an early step of the life cycle and does not require a functional macroauthophagy pathway.
a, Immunofluorescence staining of LC3B in uninfected or SARS-CoV-2-infected Huh7 cells treated with 12.5 µM of specific PI3K type 3 inhibitors for 6 hours; PI3K type 3 inhibition completely abolishes the formation of LC3-positive puncta and induces vacuoles in treated cells (bar: 25 µm). One representative picture out of four pictures taken per condition is shown. b, Immunofluorescence staining of LC3B in Huh7 cells expressing a pool of four sgRNAs targeting ATG5, ATG7, or the AAVS1 safe targeting locus. Chloroquine, an inhibitor of autophagic flux that decreases autophagosome-lysosome fusion, induces an increase in LC3-positive puncta in control cells, but fails to do so in ATG5 and ATG7 knockout cells, confirming the effective knockout of both genes (bar: 25 µm). Representative picture out of three pictures taken per condition is shown c, Time series experiment showing early stage post-receptor binding effect of PI3K type 3 inhibitor SAR405 on HCoV-229E infection. Huh7 cells were infected, treated with SAR405 at different timepoints, followed by determination of viral RNA levels at 10 hours post infection by qPCR. UDA: Urtica dioica agglutinin. Data were baseline-corrected by subtracting the signal from the first t=−0.5 timepoint for every datapoint. Combined results of six independent experiments are shown. Data are represented as mean ± s.e.m. d, Schematic representation of the wildtype SARS-CoV-2 Spike or SARS-CoV-2 spike containing two deletions sites at the N-terminus of the S1 region and near the S1/S2 cleavage site.
Extended Data Fig. 5 TMEM106B knockout decreases SARS-CoV-2 infection of different human cell lines.
a, Hep3B cells expressing control sgRNAs (AAVS1), a pool of 4 sgRNAs targeting TMEM106B and/or TMEM106B cDNA were infected with 6-fold dilutions of SARS-CoV-2 and stained with crystal violet after 3 days at 35 °C. b, Hep3B cells expressing pools of 4 sgRNAs were infected with a MOI of ~40 and stained for dsRNA at 6 hours post infection. The percentage of infected cells was determined by high content image analysis. Data were analysed by a Welch’s t test with two-tailed p-value. (n = 6 wells examined over two independent experiments). c, Different cell lines expressing control sgRNAs (targeting the safe harbour gene AAVS1) or polyclonal (Huh7 and Hep3B) or monoclonal (NCI-H1975) TMEM106B knockout cells were transduced with TMEM106B cDNA infected with 6-fold dilutions of SARS-CoV-2 and cell viability was determined after 3 days at 35 °C (MTS). AUC values were calculated and analysed by Brown-Forsythe and Welch ANOVA and Dunnet’s multiple comparison test, comparing each experimental condition to the control. For significant differences relative to the control (two-sided), p values are indicated in the figure (colored as in graph legend) (n = 6 wells examined over two independent experiments). d,e, Different cell lines expressing control sgRNAs (targeting the safe harbour gene AAVS1) or a pool of four sgRNAs targeting TMEM106B (d) and/or TMEM106B cDNA (e) were stained with a TMEM106B-specific antibody, after which TMEM106B-positive punctae were quantified by high content image analysis. Values indicate the percentage of cells containing two or more punctae (n = 3 wells), shown is the mean ± s.e.m. (bar: 40 µm).
Extended Data Fig. 6 Overexpression and knockout of TMEM106B affect SARS-CoV-2 infection of different human cell lines.
a, Huh7 cells expressing a pool of 4 sgRNAs targeting TMEM106B and/or TMEM106B cDNA were infected with SARS-CoV strain Frankfurt-1 or BetaCoV/Germany/BavPat1/2020 and stained for dsRNA 3 days after infection (n = 6 wells examined over two independent experiments). Data are expressed as % infected cells, normalized to control cells (AAVS1). Data were analysed by Brown-Forsythe and Welch ANOVA and Dunnet’s multiple comparison test P values for significant differences from the control (two-sided) are indicated. b, NCI-H2110 cells expressing pools of 4 sgRNAs were infected with SARS-CoV-2 at a MOI of ~0.2 and incubated for 7 days at 37 °C, followed by measurement of the cell viability by MTS assay (n = 6 wells examined over two independent experiments). c, A549 cells overexpressing ACE2, as well as pools of 4 sgRNAs targeting TMEM106B were infected with SARS-CoV-2 and stained for dsRNA after 6 hours (n = 6 wells examined over two independent experiments). Data are expressed as % infected cells, normalized to control cells (AAVS1). Data were analysed by a Welch t-test with two-tailed p-value. (b,c) Data are represented as mean ± s.e.m. (a,b,c) d, Wildtype A549 cells or cells expressing pools of 4 sgRNAs targeting AAVS1 or TMEM106B were stained with an ACE2-specific antibody and analyzed by flow cytometry. Left: Histogram of A549 cells overexpressing ACE2. Right: Pseudocolored dot-plots of sorted cells, showing gates used to select cells shown in the histogram. e, NCI-H1975 wt or monoclonal TMEM106B knockout cells (transduced with TMEM106B cDNA) were infected with SARS-CoV-2 at MOI 0.1. Viral RNA in supernatant was measured by qPCR at the indicated timepoints (n = 3 wells). Data were log transformed and analysed by two-way ANOVA with Geiser-Greenhouse correction and Dunnet’s multiple comparison test, comparing each experimental condition to the control. Values significantly different than the AAVS1 control cell lines are marked (***: 0.0001<p<0.001; **: 0.001<p<0.01; *: 0.01<p<0.05).
Extended Data Fig. 7 Overlap between top ranked genes of different SARS-CoV-2 genome-wide CRISPR screens.
For each screen, the 50 top ranked genes were listed and compared with the 50 top ranked genes from every other screen. Values indicate the number of overlapping genes between each set of two screens. Bold values represent the number of top 50 genes from the specific screen that were identified in at least one other screen. For Daniloski et al. a combination of the top 50 hits from the low MOI (0.01) and high MOI (0.3) screen were taken for the analysis. For the Zhu et al. screen the top 32 validated genes were taken for the analysis. For the Schneider et al. screen the top 50 genes from the 37 °C screen were included in the analysis.
Supplementary information
Rights and permissions
About this article
Cite this article
Baggen, J., Persoons, L., Vanstreels, E. et al. Genome-wide CRISPR screening identifies TMEM106B as a proviral host factor for SARS-CoV-2. Nat Genet 53, 435–444 (2021). https://doi.org/10.1038/s41588-021-00805-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-021-00805-2
This article is cited by
-
Proteomic analysis of SARS-CoV-2 particles unveils a key role of G3BP proteins in viral assembly
Nature Communications (2024)
-
Tubeimosides are pan-coronavirus and filovirus inhibitors that can block their fusion protein binding to Niemann-Pick C1
Nature Communications (2024)
-
Integrated multi-omics analyses identify anti-viral host factors and pathways controlling SARS-CoV-2 infection
Nature Communications (2024)
-
SARS-CoV-2 biology and host interactions
Nature Reviews Microbiology (2024)
-
Inflammation and neuronal gene expression changes differ in early versus late chronic traumatic encephalopathy brain
BMC Medical Genomics (2023)