Abstract
In addition to commonly associated environmental factors, genomic factors may cause cerebral palsy. We performed whole-exome sequencing of 250 parent–offspring trios, and observed enrichment of damaging de novo mutations in cerebral palsy cases. Eight genes had multiple damaging de novo mutations; of these, two (TUBA1A and CTNNB1) met genome-wide significance. We identified two novel monogenic etiologies, FBXO31 and RHOB, and showed that the RHOB mutation enhances active-state Rho effector binding while the FBXO31 mutation diminishes cyclin D levels. Candidate cerebral palsy risk genes overlapped with neurodevelopmental disorder genes. Network analyses identified enrichment of Rho GTPase, extracellular matrix, focal adhesion and cytoskeleton pathways. Cerebral palsy risk genes in enriched pathways were shown to regulate neuromotor function in a Drosophila reverse genetics screen. We estimate that 14% of cases could be attributed to an excess of damaging de novo or recessive variants. These findings provide evidence for genetically mediated dysregulation of early neuronal connectivity in cerebral palsy.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Sequencing data from University of Adelaide Robinson Research Institute (n = 154 trios) are available from the corresponding author on request, subject to human research ethics approval and patient consent. Data from PCH (n = 52 trios) are available from the corresponding author on request, subject to patient consent. Data from Zhengzhou City Children’s Hospital (n = 44 trios) are available in the CNSA of China National GeneBank DataBase repository (https://db.cngb.org/cnsa/). Source data are provided with this paper.
Change history
11 January 2021
A Correction to this paper has been published: https://doi.org/10.1038/s41588-021-00780-8.
References
Christensen, D. et al. Prevalence of cerebral palsy, co-occurring autism spectrum disorders, and motor functioning - Autism and Developmental Disabilities Monitoring Network, USA, 2008. Dev. Med. Child Neurol. 56, 59–65 (2014).
Oskoui, M., Coutinho, F., Dykeman, J., Jette, N. & Pringsheim, T. An update on the prevalence of cerebral palsy: a systematic review and meta-analysis. Dev. Med. Child Neurol. 55, 509–519 (2013).
Cans, C. Surveillance of cerebral palsy in Europe: a collaboration of cerebral palsy surveys and registers. Dev. Med. Child Neurol. 42, 816–824 (2000).
Longo, L. D. & Ashwal, S. William Osler, Sigmund Freud and the evolution of ideas concerning cerebral palsy. J. Hist. Neurosci. 2, 255–282 (1993).
Panteliadis, C., Panteliadis, P. & Vassilyadi, F. Hallmarks in the history of cerebral palsy: from antiquity to mid-20th century. Brain Dev. 35, 285–292 (2013).
Tan, S. Fault and blame, insults to the perinatal brain may be remote from time of birth. Clin. Perinatol. 41, 105–117 (2014).
Donn, S. M., Chiswick, M. L. & Fanaroff, J. M. Medico-legal implications of hypoxic–ischemic birth injury. Semin. Fetal Neonatal Med. 19, 317–321 (2014).
Korzeniewski, S. J., Slaughter, J., Lenski, M., Haak, P. & Paneth, N. The complex aetiology of cerebral palsy. Nat. Rev. Neurol. 14, 528–543 (2018).
Numata, Y. et al. Brain magnetic resonance imaging and motor and intellectual functioning in 86 patients born at term with spastic diplegia. Dev. Med. Child Neurol. 55, 167–172 (2013).
Segel, R. et al. Copy number variations in cryptogenic cerebral palsy. Neurology 84, 1660–1668 (2015).
McIntyre, S. et al. Congenital anomalies in cerebral palsy: where to from here? Dev. Med. Child Neurol. 58, 71–75 (2016).
Petterson, B., Stanley, F. & Henderson, D. Cerebral palsy in multiple births in Western Australia: genetic aspects. Am. J. Med. Genet. 37, 346–351 (1990).
Costeff, H. Estimated frequency of genetic and nongenetic causes of congenital idiopathic cerebral palsy in west Sweden. Ann. Hum. Genet. 68, 515–520 (2004).
Hallmayer, J. et al. Genetic heritability and shared environmental factors among twin pairs with autism. Arch. Gen. Psychiatry 68, 1095–1102 (2011).
Sandin, S. et al. The heritability of autism spectrum disorder. J. Am. Med. Assoc. 318, 1182–1184 (2017).
McMichael, G. et al. Rare copy number variation in cerebral palsy. Eur. J. Hum. Genet. 22, 40–45 (2014).
Oskoui, M. et al. Clinically relevant copy number variations detected in cerebral palsy. Nat. Commun. 6, 7949 (2015).
Zarrei, M. et al. De novo and rare inherited copy-number variations in the hemiplegic form of cerebral palsy. Genet. Med. 20, 172–180 (2018).
Corbett, M. A. et al. Pathogenic copy number variants that affect gene expression contribute to genomic burden in cerebral palsy. NPJ Genom. Med. 3, 33 (2018).
Takezawa, Y. et al. Genomic analysis identifies masqueraders of full-term cerebral palsy. Ann. Clin. Transl. Neurol. 5, 538–551 (2018).
Parolin Schnekenberg, R. et al. De novo point mutations in patients diagnosed with ataxic cerebral palsy. Brain 138, 1817–1832 (2015).
McMichael, G. et al. Whole-exome sequencing points to considerable genetic heterogeneity of cerebral palsy. Mol. Psychiatry 20, 176–182 (2015).
Rosenbaum, P. et al. A report: the definition and classification of cerebral palsy April 2006. Dev. Med. Child Neurol. Suppl. 109, 8–14 (2007).
Jin, S. C. et al. Contribution of rare inherited and de novo variants in 2,871 congenital heart disease probands. Nat. Genet. 49, 1593–1601 (2017).
Krumm, N. et al. Excess of rare, inherited truncating mutations in autism. Nat. Genet. 47, 582–588 (2015).
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Van der Auwera, G. A. et al. From FastQ data to high confidence variant calls: the genome analysis toolkit best practices pipeline. Curr. Protoc. Bioinformatics 43, 11.10.1–11.10.33 (2013).
Dong, C. et al. Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum. Mol. Genet. 24, 2125–2137 (2015).
Kircher, M. et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 46, 310–315 (2014).
Wei, Q. et al. A Bayesian framework for de novo mutation calling in parents–offspring trios. Bioinformatics 31, 1375–1381 (2015).
Karczewski, K. J. et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443 (2020).
Rainier, S., Sher, C., Reish, O., Thomas, D. & Fink, J. K. De novo occurrence of novel SPG3A/atlastin mutation presenting as cerebral palsy. Arch. Neurol. 63, 445–447 (2006).
Blom, N., Gammeltoft, S. & Brunak, S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 294, 1351–1362 (1999).
McNair, K. et al. A role for RhoB in synaptic plasticity and the regulation of neuronal morphology. J. Neurosci. 30, 3508–3517 (2010).
Deshaies, R. J. & Joazeiro, C. A. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78, 399–434 (2009).
Li, Y. et al. Structural basis of the phosphorylation-independent recognition of cyclin D1 by the SCFFBXO31 ubiquitin ligase. Proc. Natl Acad. Sci. USA 115, 319–324 (2018).
Vadhvani, M., Schwedhelm-Domeyer, N., Mukherjee, C. & Stegmuller, J. The centrosomal E3 ubiquitin ligase FBXO31-SCF regulates neuronal morphogenesis and migration. PLoS ONE 8, e57530 (2013).
Mir, A. et al. Truncation of the E3 ubiquitin ligase component FBXO31 causes non-syndromic autosomal recessive intellectual disability in a Pakistani family. Hum. Genet. 133, 975–984 (2014).
Lefevre, J. et al. The C terminus of tubulin, a versatile partner for cationic molecules: binding of Tau, polyamines, and calcium. J. Biol. Chem. 286, 3065–3078 (2011).
Hebebrand, M. et al. The mutational and phenotypic spectrum of TUBA1A-associated tubulinopathy. Orphanet J. Rare Dis. 14, 38 (2019).
Song, D. H. et al. CK2 phosphorylation of the armadillo repeat region of beta-catenin potentiates Wnt signaling. J. Biol. Chem. 278, 24018–24025 (2003).
Panagiotou, E. S. et al. Defects in the cell signaling mediator beta-catenin cause the retinal vascular condition FEVR. Am. J. Hum. Genet. 100, 960–968 (2017).
de Ligt, J. et al. Diagnostic exome sequencing in persons with severe intellectual disability. N. Engl. J. Med. 367, 1921–1929 (2012).
Tucci, V. et al. Dominant beta-catenin mutations cause intellectual disability with recognizable syndromic features. J. Clin. Invest. 124, 1468–1482 (2014).
Kharbanda, M. et al. Clinical features associated with CTNNB1 de novo loss of function mutations in ten individuals. Eur. J. Med. Genet. 60, 130–135 (2017).
Chen, J., Knowles, H. J., Hebert, J. L. & Hackett, B. P. Mutation of the mouse hepatocyte nuclear factor/forkhead homologue 4 gene results in an absence of cilia and random left-right asymmetry. J. Clin. Invest. 102, 1077–1082 (1998).
Orso, G. et al. Homotypic fusion of ER membranes requires the dynamin-like GTPase atlastin. Nature 460, 978–983 (2009).
Zhu, P. P., Denton, K. R., Pierson, T. M., Li, X. J. & Blackstone, C. Pharmacologic rescue of axon growth defects in a human iPSC model of hereditary spastic paraplegia SPG3A. Hum. Mol. Genet. 23, 5638–5648 (2014).
Guelly, C. et al. Targeted high-throughput sequencing identifies mutations in atlastin-1 as a cause of hereditary sensory neuropathy type I. Am. J. Hum. Genet. 88, 99–105 (2011).
Zhao, X. et al. Mutations in a newly identified GTPase gene cause autosomal dominant hereditary spastic paraplegia. Nat. Genet. 29, 326–331 (2001).
Hazan, J. et al. Spastin, a new AAA protein, is altered in the most frequent form of autosomal dominant spastic paraplegia. Nat. Genet. 23, 296–303 (1999).
Burger, J. et al. Hereditary spastic paraplegia caused by mutations in the SPG4 gene. Eur. J. Hum. Genet. 8, 771–776 (2000).
Hazan, J. et al. A fine integrated map of the SPG4 locus excludes an expanded CAG repeat in chromosome 2p-linked autosomal dominant spastic paraplegia. Genomics 60, 309–319 (1999).
de la Cruz, J., Kressler, D. & Linder, P. Unwinding RNA in Saccharomyces cerevisiae: DEAD-box proteins and related families. Trends Biochem. Sci. 24, 192–198 (1999).
Della Corte, C. M. et al. Role and targeting of anaplastic lymphoma kinase in cancer. Mol. Cancer 17, 30 (2018).
Chen, Y. et al. Oncogenic mutations of ALK kinase in neuroblastoma. Nature 455, 971–974 (2008).
Janoueix-Lerosey, I. et al. Somatic and germline activating mutations of the ALK kinase receptor in neuroblastoma. Nature 455, 967–970 (2008).
Schule, R. et al. Hereditary spastic paraplegia: clinicogenetic lessons from 608 patients. Ann. Neurol. 79, 646–658 (2016).
Parodi, L. et al. Spastic paraplegia due to SPAST mutations is modified by the underlying mutation and sex. Brain 141, 3331–3342 (2018).
Solowska, J. M., Rao, A. N. & Baas, P. W. Truncating mutations of SPAST associated with hereditary spastic paraplegia indicate greater accumulation and toxicity of the M1 isoform of spastin. Mol. Biol. Cell 28, 1728–1737 (2017).
Ji, Z. et al. Spastin interacts with CRMP5 to promote neurite outgrowth by controlling the microtubule dynamics. Dev. Neurobiol. 78, 1191–1205 (2018).
Gao, Y. et al. Atlastin-1 regulates dendritic morphogenesis in mouse cerebral cortex. Neurosci. Res. 77, 137–142 (2013).
Romeo, D. M. et al. Sex differences in cerebral palsy on neuromotor outcome: a critical review. Dev. Med. Child Neurol. 58, 809–813 (2016).
Reid, S. M., Meehan, E. M., Arnup, S. J. & Reddihough, D. S. Intellectual disability in cerebral palsy: a population-based retrospective study. Dev. Med. Child Neurol. 60, 687–694 (2018).
Pinero, J. et al. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 45, D833–D839 (2017).
Szklarczyk, D. et al. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613 (2019).
Al-Mubarak, B. et al. Whole exome sequencing reveals inherited and de novo variants in autism spectrum disorder: a trio study from Saudi families. Sci. Rep. 7, 5679 (2017).
Giacopuzzi, E. et al. Exome sequencing in schizophrenic patients with high levels of homozygosity identifies novel and extremely rare mutations in the GABA/glutamatergic pathways. PLoS ONE 12, e0182778 (2017).
Huang da, W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009).
Liberzon, A. et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 (2015).
Mi, H. et al. Protocol Update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0). Nat. Protoc. 14, 703–721 (2019).
Fang, H. & Gough, J. DcGO: database of domain-centric ontologies on functions, phenotypes, diseases and more. Nucleic Acids Res. 41, D536–D544 (2013).
Novarino, G. et al. Exome sequencing links corticospinal motor neuron disease to common neurodegenerative disorders. Science 343, 506–511 (2014).
Stessman, H. A. et al. Targeted sequencing identifies 91 neurodevelopmental-disorder risk genes with autism and developmental-disability biases. Nat. Genet. 49, 515–526 (2017).
Estes, P. S. et al. Wild-type and A315T mutant TDP-43 exert differential neurotoxicity in a Drosophila model of ALS. Hum. Mol. Genet. 20, 2308–2321 (2011).
Madabattula, S. T. et al. Quantitative analysis of climbing defects in a Drosophila model of neurodegenerative disorders. J. Vis. Exp. https://doi.org/10.3791/52741 (2015).
Kim, M. et al. Mutation in ATG5 reduces autophagy and leads to ataxia with developmental delay. eLife 5, e12245 (2016).
Aleman-Meza, B., Loeza-Cabrera, M., Pena-Ramos, O., Stern, M. & Zhong, W. High-content behavioral profiling reveals neuronal genetic network modulating Drosophila larval locomotor program. BMC Genet. 18, 40 (2017).
Hemminki, K., Li, X., Sundquist, K. & Sundquist, J. High familial risks for cerebral palsy implicate partial heritable aetiology. Paediatr. Perinat. Epidemiol. 21, 235–241 (2007).
MacLennan, A. H. et al. Cerebral palsy and genomics: an international consortium. Dev. Med. Child Neurol. 60, 209–210 (2018).
Himmelmann, K. & Uvebrant, P. The panorama of cerebral palsy in Sweden part XII shows that patterns changed in the birth years 2007–2010. Acta Paediatr. 107, 462–468 (2018).
van Eyk, C. L. et al. Analysis of 182 cerebral palsy transcriptomes points to dysregulation of trophic signalling pathways and overlap with autism. Transl. Psychiatry 8, 88 (2018).
Martinelli, S. et al. Functional dysregulation of CDC42 causes diverse developmental phenotypes. Am. J. Hum. Genet. 102, 309–320 (2018).
Englander, Z. A. et al. Brain structural connectivity increases concurrent with functional improvement: evidence from diffusion tensor MRI in children with cerebral palsy during therapy. Neuroimage Clin. 7, 315–324 (2015).
Loubet, D. et al. Neuritogenesis: the prion protein controls beta1 integrin signaling activity. FASEB J. 26, 678–690 (2012).
Colombo, S. et al. G protein-coupled potassium channels implicated in mouse and cellular models of GNB1 Encephalopathy. Preprint at bioRxiv https://doi.org/10.1101/697235 (2019).
Pipo-Deveza, J. et al. Rationale for dopa-responsive CTNNB1/β-catenin deficient dystonia. Mov. Disord. 33, 656–657 (2018).
Akizu, N. et al. AMPD2 regulates GTP synthesis and is mutated in a potentially treatable neurodegenerative brainstem disorder. Cell 154, 505–517 (2013).
van Eyk, C. L. et al. Targeted resequencing identifies genes with recurrent variation in cerebral palsy. NPJ Genom. Med. 4, 27 (2019).
Miller, S. P., Shevell, M. I., Patenaude, Y. & O’Gorman, A. M. Neuromotor spectrum of periventricular leukomalacia in children born at term. Pediatr. Neurol. 23, 155–159 (2000).
Li, H. & Durbin, R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26, 589–595 (2010).
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010).
Lek, M. et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 (2016).
1000 Genomes Project Consortium A global reference for human genetic variation. Nature 526, 68–74 (2015).
Ware, J. S., Samocha, K. E., Homsy, J. & Daly, M. J. Interpreting de novo variation in human disease using denovolyzeR. Curr. Protoc. Hum. Genet. 87, 7.25.1–7.25.15 (2015).
Homsy, J. et al. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science 350, 1262–1266 (2015).
Huang da, W., Sherman, B. T. & Lempicki, R. A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 (2009).
Mi, H., Muruganujan, A., Ebert, D., Huang, X. & Thomas, P. D. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 47, D419–D426 (2019).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Acknowledgements
We gratefully acknowledge the support of the patients and families who have graciously and patiently supported this work from its inception. Without their partnership, these studies would not have been possible. We acknowledge the support of the clinicians who generously provided their expertise in support of this study, including M.-C. Waugh, M. Axt and V. Roberts of the Children’s Hospital Westmead; K. Lowe of Sydney Children’s Hospital; R. Russo, J. Rice and A. Tidemann of the Women’s and Children’s Hospital, Adelaide; T. Carroll and L. Copeland of the Lady Cilento Children’s Hospital, Brisbane; and J. Valentine of Perth Children’s Hospital. We appreciate the collaboration of S. Knoblach and E. Hoffman (Children’s National Medical Center). This work was supported in part by the Cerebral Palsy Alliance Research Foundation (M.C.K.), the Yale-NIH Center for Mendelian Genomics (U54 HG006504-01), Doris Duke Charitable Foundation CSDA 2014112 (M.C.K.), the Scott Family Foundation (M.C.K.), Cure CP (M.C.K.), NHMRC grant 1099163 (A.H.M., C.L.v.E., J.G. and M.A.C.), NHMRC Senior Principal Research Fellowship 1155224 (J.G.), Channel 7 Children’s Research Foundation (J.G.), a Cerebral Palsy Alliance Research Foundation Career Development Award (M.A.C.), the Tenix Foundation (A.H.M., J.G., C.L.v.E. and M.A.C.), the National Natural Science Foundation of China (U1604165, X.W.), Henan Key Research Program of China (171100310200, C. Zhu), VINNOVA (2015-04780, C. Zhu), the James Hudson Brown–Alexander Brown Coxe Postdoctoral Fellowship at the Yale University School of Medicine (S.C.J.), an American Heart Association Postdoctoral Fellowship (18POST34060008 to S.C.J.), the NIH K99/R00 Pathway to Independence Award (R00HL143036-02 to S.C.J.) and NIH grants R01NS091299 (D.C.Z.) and NIH R01NS106298 (M.C.K.).
Author information
Authors and Affiliations
Contributions
K.B., S.P.-L., Q.X., C. Zhu, R.P.L., A.H.M., J.G. and M.C.K. contributed to study design, data interpretation and oversight. B.Y.N., J.G.B., K.H., C. Zhou, D.Z., B.Z., B.K., S.W., J.B., S.P., J.B.V., J.B.-H., A.P., M.C.F., L.X., Y.X., M.C., K.R., F.M., Y.W., J.L.W., L.R., J.S.C., A.F., A.E.L., J.P.P., T.F., S.J.M., K.E.C., S.M.R., D.S.R., Q.S., C.G., Y.A.W., N.B., I.N., S.C.M., X.W., D.J.A., J.H. and M.C.K. provided cohort ascertainment, recruitment and phenotypic characterization. K.B., C.C., A.E., J.L., C.L.v.E., H.M., S.M.M., I.R.T., F.L.-G., Y.A.W., B.S.G., J.Z., D.L.W., M.S.B.F., C. Zhou and M.A.C. performed exome sequencing production and validation. S.B., S.C.J., M.A.C., M.C.S., X.Z., J.R.K. and A.H.S. performed WES analysis. A.E., H.M., J.L., B.S.G. and S.P.-L. performed RHOB validation. S.M.N., S.P.-L., S.P., J.B.V., D.D. and S.A.L. performed FBXO31 validation. S.A.L., S.V. and D.C.Z. performed Drosophila locomotor experiments. S.C.J., S.A.L., S.B., S.S., B.L., Q.L., M.C.S. and X.Z. conducted statistical analysis. S.H. performed biophysical simulation for RHOB and FBXO31. S.C.J., S.A.L., J.G., Q.L., S.P.-L., R.P.L., A.H.M., S.M., B.Y.N., M.C.S., X.Z., C.L.v.E., X.W., Q.X., C. Zhu and M.C.K. wrote and reviewed the manuscript. K.B., R.P.L., Q.X., C. Zhu, A.H.M., J.G., S.P.-L. and M.C.K. acquired funding and supervised the project and were considered co-senior authors. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Brain MRI features of idiopathic cerebral palsy.
F050: bilateral periventricular leukomalacia; F055: right sided porencephaly; F057: normal (equivocal putaminal rim hyperintensity); F063: mildly globally diminished cerebral volume; F066: normal; F068, bilateral mild periventricular leukomalacia, white matter thinning and colpocephaly; F069: diminished cortical more than cerebellar volumes; F074: normal; F076: ex vacuo ventriculomegaly; bilateral periventricular leukomalacia, and bilateral perisylvian pachygyria; F077: mild periventricular leukomalacia; F082: scattered subcortical T2 hyperintensities; F084: normal; F085: colpocephaly, thinning of periventricular white matter, hypoplastic corpus callosum, diminished left cerebellar hemispheric volume; F093: normal; F124: normal; F162: normal; F217: equivocal ex vacuo ventriculomegaly; F218: normal; F300: bilateral periventricular leukomalacia with thin corpus callosum; F306: scattered bilateral subcortical punctate T2/FLAIR hyperintensities; F309: simplified gyral pattern; F311: normal; F312: normal; F313: normal; F342: diminished cortical volume, thinning and T2/FLAIR signal hyperintensity of periventricular white matter, thin corpus callosum; F356: bilateral perisylvian polymicrogyria; F357: thin corpus callosum; F377: equivocally simplified gyri with ‘open opercula’; F383: bilateral occipital horn heterotopias; F385: hydrocephalus and periventricular leukomalacia; F393: periventricular leukomalacia; F433: normal; F439: increased frontotemporal extra-axial fluid spaces and thin corpus callosum; F444: normal (equivocally thickened corpus callosum); F468: slight ex vacuo ventriculomegaly; F470: equivocally diminished cortical volume; F606: bilateral perislyvian pachygyria; F609: bi hemispheric periventricular leukomalacia; F617: ex vacuo ventriculomegaly; F623: dysplastic corpus callosum, bitemporal diminished cortical volumes; F629: thin corpus callosum, colpocephaly, with periventricular leukomalacia; F648: periventricular leukomalacia; F658: right sided encephalomalacia affecting putamen and thalamus.
Extended Data Fig. 2 De novo mutation rate closely approximates Poisson distribution in cases and controls.
Observed number of de novo mutations per subject (bars) compared to the numbers expected (line) from the Poisson distribution in the case (red) and control (blue) cohorts. Here, ‘P’ denotes chi-squared P-value.
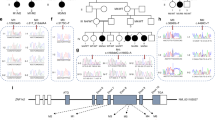
Extended Data Fig. 3 De novo mutation in TUBA1A encoding α-tubulin.
a, TUBA1A functional domains schematic with locations of previously-described pathogenic variants (red) compared to those from this work (black). b, Phylogenetic conservation of reference amino acid at each mutated position described in this work. c, Sanger-verified mutated base (red arrow) with the corresponding reference bases. d, MRI of the brain (F356) demonstrates evidence of bilateral perisylvian pachygyria (blue arrows). Conserved Domain Annotations: TNBDL (AA 1-244) as IPro36525; SD (AA 418-451) annotated as per39.
Extended Data Fig. 4 De novo mutations in CTNNB1 encoding β-catenin.
a, CTNNB1 functional domain with location of previously reported pathogenic variants (red) and those identified in this work (black). (Given the loss-of-function nature of the identified variants, phylogenetic alignments were not performed; however, 100% identify is seen at these loci (p.E54, p.F99, and p.R449) in primates). b, Sanger-verified mutated base (red arrow) with corresponding reference bases. c, Brain MRI (F066) was unremarkable. Conserved Domain Annotations: ARM, Armadillo/beta-catenin-like repeats from UniProtKB/Swiss-Prot (P35222.1); SCRIB, interaction with SCRIB (AA 772-781, by similarity, experimental evidence); BCL9, interaction with BCL9 (AA 156-178, by similarity, experimental evidence); VCL, interaction with VCL (AA 2-23, by similarity, experimental evidence).
Extended Data Fig. 5 De novo mutations in ATL1 encoding atlastin-1.
a, ATL1 functional domain with location of previously reported variants (red) as well as those identified in this work (black). b, Phylogenetic conservation of reference amino acid at each affected position. c, Sanger-verified mutated base (red arrow) with the corresponding reference bases. d, Brain MRI images from F050 and F609 demonstrate mild periventricular T2 hyperintensity (blue arrows). Conserved Domain Annotations: GBP (AA 43-314) as pfam02263; Membrane localization domain (AA 448-558) from UniProtKB (Q8WXF7.1).
Extended Data Fig. 6 De novo mutations in SPAST encoding spastin.
a, SPAST functional domains with location of CP-associated damaging variants identified in this study (black); 277 pathological mutations58 have previously been identified in SPAST with the majority (82%) located within the conserved domains (red). b, Phylogenetic conservation of wild-type amino acid at each mutated position. c, Sanger-verified mutated base indicated by red arrow with corresponding reference bases. d, Brain MRI (F082) showed mild subcortical T2 hyperintensities (blue arrows). Conserved Domain Annotations: MIT (AA 116-196) as CDD:239142; Microtubule Binding domain (AA 270-328) from UniProtKB/Swiss-Prot (Q9UBP0.1); ATPase AAA Core and Lid domains (378-567) from IPR003959 and IPR041569, respectively.
Extended Data Fig. 7 De novo mutations in DHX32 encoding the DEAH box polypeptide 32.
a, DHX32 functional domains with location of CP-associated damaging variants from this work (black). Germline DHX32 variants have not been previously associated with human disease although somatic variants (>40) have been associated with variants cancers (COSMIC). b, Phylogenetic conservation of wild-type amino acid at each mutated position. c, Sanger-verified mutated base indicated by red arrow with corresponding reference bases. d, Brain MRI (F063) showed diffusely diminished cortical volume. Conserved Domain Annotations: Helicase and DEAD domains overlap (72-378 and 146-403) from IPR014001 and cd17912, respectively; HA2 domain (AA 458-547) as IPR007502; Helicase associated domain of unknown function (AA 616-696) from IPR011709.
Extended Data Fig. 8 De novo mutations in ALK encoding the anaplastic lymphoma kinase.
a, ALK functional domain with location of previously reported pathogenic variants associated with susceptibility to neuroblastoma (OMIM# 613014) (red) as well as CP-associated damaging variants identified in this work (black). b, Phylogenetic conservation of wild-type amino acid at each mutated position. c, Sanger-verified mutated base indicated by red arrow with corresponding reference bases. d, Brain MRI (F306) demonstrates punctate subcortical T2 hyperintensities of both hemispheres. Conserved Domain Annotations: Signal Peptide (AA 1-18) by SignalP 4.0; MAM (AA 266-427, 480-636) as pfam #00629; LDLa (AA 441-467) as smart#00192; Fxa (AA 987-1021) as pfam#14670; PTKc ALK LTK (AA 1109-1385) as CDD#05036.
Extended Data Fig. 9 Additional locomotor phenotypes of loss of function mutations in Drosophila orthologs of candidate cerebral palsy risk genes.
Drosophila mutant and control genotypes are shown in Supplementary Table 9. a, Turning time, a measure of coordinated movements, is increased in larva with mutations in AKT3 and PNPLA7 orthologs, but not in MAP2K4. b-o, Distance threshold assay examining negative geotaxis climbing defects in for 14 day-old adult flies with mutations in orthologs of AGAP1 (b), AKT3 (c), ANKS1A (d), ARHGEF17 (e), DIAPH2 (f), HSPG2 (g), KIDINS220 (h), MAP2K4 (i), MPP1 (j), PNPLA7 (k), PRICKLE1 (l), SYNGAP1 (m), TBC1D17 (n), and TENM1 (o). Impairments in the climbing assay was detected for males with mutations in AKT3 and PRICKLE1 (c,l) and for both sexes with mutations in MAP2K4 and MPP1 (i,j) orthologs. Climbing phenotype mapped to gene using deficiency chromosome for AGAP1 (b), but did not map for TENM1 (o). There was no locomotor impairment in the two negative control genotypes, ARHGEF15 and ANKS1A, where the patient variant did not pass our deleteriousness filters (d). For larval turning, box indicates 75th and 25th percentile with median line; whiskers indicate 10th and 90th percentile (n = 50 larvae). Locomotor curve represents average of all trials and bars indicate standard error (n = 10-21 trials). Statistics between larval turning times determined using unpaired 2-tailed t-test. Locomotor curves considered to be significantly different from each other if P < 0.05 for Kolomogrov-Smirnov test in addition to a significant difference at one or more time bins by Mann-Whitney rank sum 2-tailed test. *P < 0.05, ****P < 1 ×10−6. Exact genotypes, n, and P values are provided in Supplementary Table 9.
Extended Data Fig. 10 Cerebral palsy gene discovery projections.
a, Estimation of the number of cerebral palsy risk genes via de novo mechanism. Monte Carlo simulation performed was performed based on observed damaging de novo mutations in 3,049 loss-of-function intolerant genes (pLI ≥ 0.9 in gnomAD (v2.1.1)) using 20,000 iterations. We estimate that the number of risk genes via de novo events to be ~75 (95% confidence interval = (26.5, 123.5)). b, Estimation of the number of recurrent genes. The number of trios and the number of genes with more than one damaging de novo mutation are specified on the x and y-axis, respectively. We modeled the expected rate of damaging de novo mutations given an increasing sample size. A total of 10,000 iterations were performed to estimate the number of genes with more than one damaging de novo mutations taking into account of the damaging de novo mutation probability. WES of 2,500 and 7,500 trios are expected to yield a 65.3% and 91.8% saturation rate, respectively, for all cerebral palsy risk genes.
Supplementary information
Supplementary Information
Supplementary Note
Supplementary Data
Supplementary Datasets 1–15
Supplementary Tables
Supplementary Tables 1–9
Source data
Source Data Fig. 1
Unprocessed western blots for RHOB.
Source Data Fig. 2
Unprocessed western blots for FBOX31.
Rights and permissions
About this article
Cite this article
Jin, S.C., Lewis, S.A., Bakhtiari, S. et al. Mutations disrupting neuritogenesis genes confer risk for cerebral palsy. Nat Genet 52, 1046–1056 (2020). https://doi.org/10.1038/s41588-020-0695-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-020-0695-1
This article is cited by
-
Comprehensive whole-genome sequence analyses provide insights into the genomic architecture of cerebral palsy
Nature Genetics (2024)
-
Key role of Rho GTPases in motor disorders associated with neurodevelopmental pathologies
Molecular Psychiatry (2023)
-
Anterior segment indices in mentally retarded children
Scientific Reports (2023)
-
Redefining cerebral palsies as a diverse group of neurodevelopmental disorders with genetic aetiology
Nature Reviews Neurology (2023)
-
Neurodevelopmental disorders, like cancer, are connected to impaired chromatin remodelers, PI3K/mTOR, and PAK1-regulated MAPK
Biophysical Reviews (2023)