Abstract
Pachytene PIWI-interacting RNAs (piRNAs), which comprise >80% of small RNAs in the adult mouse testis, have been proposed to bind and regulate target RNAs like microRNAs, cleave targets like short interfering RNAs or lack biological function altogether. Although piRNA pathway protein mutants are male sterile, no biological function has been identified for any mammalian piRNA-producing locus. Here, we report that males lacking piRNAs from a conserved mouse pachytene piRNA locus on chromosome 6 (pi6) produce sperm with defects in capacitation and egg fertilization. Moreover, heterozygous embryos sired by pi6−/− fathers show reduced viability in utero. Molecular analyses suggest that pi6 piRNAs repress gene expression by cleaving messenger RNAs encoding proteins required for sperm function. pi6 also participates in a network of piRNA–piRNA precursor interactions that initiate piRNA production from a second piRNA locus on chromosome 10, as well as pi6 itself. Our data establish a direct role for pachytene piRNAs in spermiogenesis and embryo viability.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
All sequencing data are available through the National Center for Biotechnology Information Sequence Read Archive using accession number PRJNA634688. Source data are provided with this paper.
Code availability
The code used for identifying piRNA-directed cleavage sites is available at https://github.com/weng-lab/GTBuster. All other codes used in this study are described in the Methods and Nature Research Reporting Summary. Source data are provided with this paper.
References
Aravin, A. A. et al. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol. Cell 31, 785–799 (2008).
Lewis, S. H. et al. Pan-arthropod analysis reveals somatic piRNAs as an ancestral defence against transposable elements. Nat. Ecol. Evol. 2, 174–181 (2018).
Batista, P. J. et al. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol. Cell 31, 67–78 (2008).
Das, P. P. et al. Piwi and piRNAs act upstream of an endogenous siRNA pathway to suppress Tc3 transposon mobility in the Caenorhabditis elegans germline. Mol. Cell 31, 79–90 (2008).
Houwing, S. et al. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in zebrafish. Cell 129, 69–82 (2007).
Robine, N. et al. A broadly conserved pathway generates 3′ UTR-directed primary piRNAs. Curr. Biol. 19, 2066–2076 (2009).
Ozata, D. M. et al. Evolutionarily conserved pachytene piRNA loci are highly divergent among modern humans. Nat. Ecol. Evol. 4, 156–168 (2019).
Chirn, G. W. et al. Conserved piRNA expression from a distinct set of piRNA cluster loci in eutherian mammals. PLoS Genet. 11, e1005652 (2015).
Girard, A., Sachidanandam, R., Hannon, G. J. & Carmell, M. A. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 442, 199–202 (2006).
Li, X. Z. et al. An ancient transcription factor initiates the burst of piRNA production during early meiosis in mouse testes. Mol. Cell 50, 67–81 (2013).
Lau, N. C. et al. Characterization of the piRNA complex from rat testes. Science 313, 363–367 (2006).
Grivna, S. T., Beyret, E., Wang, Z. & Lin, H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 20, 1709–1714 (2006).
Ro, S. et al. Cloning and expression profiling of testis-expressed piRNA-like RNAs. RNA 13, 1693–1702 (2007).
Bolcun-Filas, E. et al. A-MYB (MYBL1) transcription factor is a master regulator of male meiosis. Development 138, 3319–3330 (2011).
Deng, W. & Lin, H. Miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev. Cell 2, 819–830 (2002).
Reuter, M. et al. Miwi catalysis is required for piRNA amplification-independent LINE1 transposon silencing. Nature 480, 264–267 (2011).
Zheng, K. & Wang, P. J. Blockade of pachytene piRNA biogenesis reveals a novel requirement for maintaining post-meiotic germline genome integrity. PLoS Genet. 8, e1003038 (2012).
Wasik, K. A. et al. RNF17 blocks promiscuous activity of PIWI proteins in mouse testes. Genes Dev. 29, 1403–1415 (2015).
Castañeda, J. et al. Reduced pachytene piRNAs and translation underlie spermiogenic arrest in Maelstrom mutant mice. EMBO J. 33, 1999–2019 (2014).
Homolka, D. et al. PIWI slicing and RNA elements in precursors instruct directional primary piRNA biogenesis. Cell Rep. 12, 418–428 (2015).
Xu, M. et al. Mice deficient for a small cluster of Piwi-interacting RNAs implicate Piwi-interacting RNAs in transposon control. Biol. Reprod. 79, 51–57 (2008).
Goh, W. S. et al. piRNA-directed cleavage of meiotic transcripts regulates spermatogenesis. Genes Dev. 29, 1032–1044 (2015).
Zhang, P. et al. MIWI and piRNA-mediated cleavage of messenger RNAs in mouse testes. Cell Res. 25, 193–207 (2015).
Gou, L. T. et al. Pachytene piRNAs instruct massive mRNA elimination during late spermiogenesis. Cell Res. 24, 680–700 (2014).
Vourekas, A. et al. Mili and Miwi target RNA repertoire reveals piRNA biogenesis and function of Miwi in spermiogenesis. Nat. Struct. Mol. Biol. 19, 773–781 (2012).
Palumbo, G., Bonaccorsi, S., Robbins, L. G. & Pimpinelli, S. Genetic analysis of Stellate elements of Drosophila melanogaster. Genetics 138, 1181–1197 (1994).
Mével-Ninio, M., Pelisson, A., Kinder, J., Campos, A. R. & Bucheton, A. The flamenco locus controls the gypsy and ZAM retroviruses and is required for Drosophila oogenesis. Genetics 175, 1615–1624 (2007).
Bozzetti, M. P. et al. The Ste locus, a component of the parasitic cry-Ste system of Drosophila melanogaster, encodes a protein that forms crystals in primary spermatocytes and mimics properties of the beta subunit of casein kinase 2. Proc. Natl Acad. Sci. USA 92, 6067–6071 (1995).
Livak, K. J. Detailed structure of the Drosophila melanogaster stellate genes and their transcripts. Genetics 124, 303–316 (1990).
Livak, K. J. Organization and mapping of a sequence on the Drosophila melanogaster X and Y chromosomes that is transcribed during spermatogenesis. Genetics 107, 611–634 (1984).
Robert, V., Prud’homme, N., Kim, A., Bucheton, A. & Pélisson, A. Characterization of the flamenco region of the Drosophila melanogaster genome. Genetics 158, 701–713 (2001).
Prud’homme, N., Gans, M., Masson, M., Terzian, C. & Bucheton, A. flamenco, a gene controlling the gypsy retrovirus of Drosophila melanogaster. Genetics 139, 697–711 (1995).
Pélisson, A. et al. Gypsy transposition correlates with the production of a retroviral envelope-like protein under the tissue-specific control of the Drosophila flamenco gene. EMBO J. 13, 4401–4411 (1994).
Aravin, A. A. et al. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr. Biol. 11, 1017–1027 (2001).
Aravin, A. A. et al. The small RNA profile during Drosophila melanogaster development. Dev. Cell 5, 337–350 (2003).
Saito, K. et al. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature 461, 1296–1299 (2009).
Brennecke, J. et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128, 1089–1103 (2007).
Bourc’his, D. & Bestor, T. H. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature 431, 96–99 (2004).
Ahmadi, A. & Ng, S. C. Fertilizing ability of DNA-damaged spermatozoa. J. Exp. Zool. 284, 696–704 (1999).
Morris, I. D., Ilott, S., Dixon, L. & Brison, D. R. The spectrum of DNA damage in human sperm assessed by single cell gel electrophoresis (Comet assay) and its relationship to fertilization and embryo development. Hum. Reprod. 17, 990–998 (2002).
Lewis, S. E. & Aitken, R. J. DNA damage to spermatozoa has impacts on fertilization and pregnancy. Cell Tissue Res. 322, 33–41 (2005).
Aravin, A. et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442, 203–207 (2006).
Kuretake, S., Kimura, Y., Hoshi, K. & Yanagimachi, R. Fertilization and development of mouse oocytes injected with isolated sperm heads. Biol. Reprod. 55, 789–795 (1996).
de Lamirande, E., Leclerc, P. & Gagnon, C. Capacitation as a regulatory event that primes spermatozoa for the acrosome reaction and fertilization. Mol. Hum. Reprod. 3, 175–194 (1997).
Florman, H. M. & Storey, B. T. Mouse gamete interactions: the zona pellucida is the site of the acrosome reaction leading to fertilization in vitro. Dev. Biol. 91, 121–130 (1982).
Jin, M. et al. Most fertilizing mouse spermatozoa begin their acrosome reaction before contact with the zona pellucida during in vitro fertilization. Proc. Natl Acad. Sci. USA 108, 4892–4896 (2011).
Stauss, C. R., Votta, T. J. & Suarez, S. S. Sperm motility hyperactivation facilitates penetration of the hamster zona pellucida. Biol. Reprod. 53, 1280–1285 (1995).
Suarez, S. S., Katz, D. F., Owen, D. H., Andrew, J. B. & Powell, R. L. Evidence for the function of hyperactivated motility in sperm. Biol. Reprod. 44, 375–381 (1991).
Qi, H. et al. All four CatSper ion channel proteins are required for male fertility and sperm cell hyperactivated motility. Proc. Natl Acad. Sci. USA 104, 1219–1223 (2007).
Quill, T. A. et al. Hyperactivated sperm motility driven by CatSper2 is required for fertilization. Proc. Natl Acad. Sci. USA 100, 14869–14874 (2003).
Mortimer, S. T. CASA—practical aspects. J. Androl. 21, 515–524 (2000).
Goodson, S. G., Zhang, Z., Tsuruta, J. K., Wang, W. & O’Brien, D. A. Classification of mouse sperm motility patterns using an automated multiclass support vector machines model. Biol. Reprod. 84, 1207–1215 (2011).
González-Jara, P. et al. Optimization of the balance between effort and yield in unilateral surgical transfer of mouse embryos. Lab. Anim. 51, 622–628 (2017).
Sun-Wada, G. H. et al. A proton pump ATPase with testis-specific E1-subunit isoform required for acrosome acidification. J. Biol. Chem. 277, 18098–18105 (2002).
Huang, T. T. et al. pH and protease control of acrosomal content stasis and release during the guinea pig sperm acrosome reaction. Biol. Reprod. 32, 451–462 (1985).
Son, S. W. et al. Prognostic significance and function of the vacuolar H+-ATPase subunit V1E1 in esophageal squamous cell carcinoma. Oncotarget 7, 49334–49348 (2016).
Chung, J. J. et al. CatSperζ regulates the structural continuity of sperm Ca2+ signaling domains and is required for normal fertility. eLife 6, e23082 (2017).
Brown, S. G. et al. Homozygous in-frame deletion in CATSPERE in a man producing spermatozoa with loss of CatSper function and compromised fertilizing capacity. Hum. Reprod. 33, 1812–1816 (2018).
Gunawardane, L. S. et al. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 315, 1587–1590 (2007).
Mohn, F., Handler, D. & Brennecke, J. piRNA-guided slicing specifies transcripts for Zucchini-dependent, phased piRNA biogenesis. Science 348, 812–817 (2015).
Han, B. W., Wang, W., Li, C., Weng, Z. & Zamore, P. D. Noncoding RNA piRNA-guided transposon cleavage initiates Zucchini-dependent, phased piRNA production. Science 348, 817–821 (2015).
Gainetdinov, I., Colpan, C., Arif, A., Cecchini, K. & Zamore, P. D. A single mechanism of biogenesis, initiated and directed by PIWI proteins, explains piRNA production in most animals. Mol. Cell 71, 775–790.e5 (2018).
Post, C., Clark, J. P., Sytnikova, Y. A., Chirn, G. W. & Lau, N. C. The capacity of target silencing by Drosophila PIWI and piRNAs. RNA 20, 1977–1986 (2014).
Kiuchi, T. et al. A single female-specific piRNA is the primary determiner of sex in the silkworm. Nature 509, 633–636 (2014).
Bartel, D. P. Metazoan microRNAs. Cell 173, 20–51 (2018).
Becker, W. R. et al. High-throughput analysis reveals rules for target RNA binding and cleavage by AGO2. Mol. Cell 75, 741–755.e11 (2019).
Wee, L. M., Flores-Jasso, C. F., Salomon, W. E. & Zamore, P. D. Argonaute divides its RNA guide into domains with distinct functions and RNA-binding properties. Cell 151, 1055–1067 (2012).
Bartel, D. P. MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233 (2009).
Borini, A. et al. Sperm DNA fragmentation: paternal effect on early post-implantation embryo development in ART. Hum. Reprod. 21, 2876–2881 (2006).
Sakkas, D. et al. Sperm nuclear DNA damage and altered chromatin structure: effect on fertilization and embryo development. Hum. Reprod. 13, 11–19 (1998).
Peaston, A. E. et al. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev. Cell 7, 597–606 (2004).
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013).
Truett, G. E. et al. Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT). Biotechniques 29, 52–54 (2000).
Holloway, J. K., Sun, X., Yokoo, R., Villeneuve, A. M. & Cohen, P. E. Mammalian CNTD1 is critical for meiotic crossover maturation and deselection of excess precrossover sites. J. Cell Biol. 205, 633–641 (2014).
Cole, F. et al. Mouse tetrad analysis provides insights into recombination mechanisms and hotspot evolutionary dynamics. Nat. Genet. 46, 1072–1080 (2014).
Nagy, A., Gertsenstein, M. V. K. & Behringer, R. Manipulating the Mouse Embryo: A Laboratory Manual (Cold Spring Harbor Laboratory Press, 2003).
Yanagimachi, R., Yanagimachi, H. & Rogers, B. J. The use of zona-free animal ova as a test-system for the assessment of the fertilizing capacity of human spermatozoa. Biol. Reprod. 15, 471–476 (1976).
Johnson, A., Smith, R. G., Bassham, B., Lipshultz, L. I. & Lamb, D. J. The microsperm penetration assay: development of a sperm penetration assay suitable for oligospermic males. Fertil. Steril. 56, 528–534 (1991).
Talbot, P., Summers, R. G., Hylander, B. L., Keough, E. M. & Franklin, L. E. The role of calcium in the acrosome reaction: an analysis using ionophore A23187. J. Exp. Zool. 198, 383–392 (1976).
Osman, R. A., Andria, M. L., Jones, A. D. & Meizel, S. Steroid induced exocytosis: the human sperm acrosome reaction. Biochem. Biophys. Res. Commun. 160, 828–833 (1989).
Arnoult, C., Zeng, Y. & Florman, H. M. ZP3-dependent activation of sperm cation channels regulates acrosomal secretion during mammalian fertilization. J. Cell Biol. 134, 637–645 (1996).
Tateno, H. et al. Ca2+ ionophore A23187 can make mouse spermatozoa capable of fertilizing in vitro without activation of cAMP-dependent phosphorylation pathways. Proc. Natl Acad. Sci. USA 110, 18543–18548 (2013).
Mortimer, D., Curtis, E. F. & Miller, R. G. Specific labelling by peanut agglutinin of the outer acrosomal membrane of the human spermatozoon. J. Reprod. Fertil. 81, 127–135 (1987).
Fu, Y., Wu, P. H., Beane, T., Zamore, P. D. & Weng, Z. Elimination of PCR duplicates in RNA-seq and small RNA-seq using unique molecular identifiers. BMC Genomics 19, 531 (2018).
Han, B. W., Wang, W., Zamore, P. D. & Weng, Z. piPipes: a set of pipelines for piRNA and transposon analysis via small RNA-seq, RNA-seq, degradome- and CAGE-seq, ChIP-seq and genomic DNA sequencing. Bioinformatics 31, 593–595 (2015).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Addo-Quaye, C., Eshoo, T. W., Bartel, D. P. & Axtell, M. J. Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr. Biol. 18, 758–762 (2008).
Wang, W. et al. Slicing and binding by Ago3 or Aub trigger Piwi-bound piRNA production by distinct mechanisms. Mol. Cell 59, 819–830 (2015).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Acknowledgements
We thank P. Cohen, K. Grive and E. Crate at Cornell University for generously sharing protocols and advice on germ cell sorting and meiotic chromosome studies; H. Florman, P. Visconti and M. Gervasi for sharing protocols and advice on sperm studies; the UMMS Transgenic Animal Modeling Core for advice on fertility test and embryo phenotype; the UMMS FACS core for advice on and help with germ cell sorting; the UMMS EM Core (supported by National Center for Research Resources Award SI0OD021580) for advice on and help with sperm transmission electron microscopy; and members of our laboratories for critical comments on the manuscript. This work was supported in part by National Institutes of Health grants GM62862 to P.D.Z. and P01HD078253 to P.D.Z. and Z.W.
Author information
Authors and Affiliations
Contributions
P.-H.W., K.C., Y.F., Z.W. and P.D.Z. conceived and designed the experiments. P.-H.W., K.C., D.M.Ö., A.A. and C.C. performed the experiments. Y.F., T.Y., I.G. and P.-H.W. analyzed the sequencing data. P.-H.W., Y.F. and P.D.Z. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
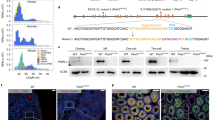
Extended Data Fig. 1 Confirmation of mutant founder genotypes.
a, Genotyping of mutant founders by PCR. Mutant founders were generated by injecting sgRNAs and Cas9 mRNAs into C57BL/6 one-cell zygotes, which were transferred to surrogate mothers and screened after birth. Gel images were cropped for clarity (see also Source Data). Genomic sequences of pi6 promoter region in pi6em1 (b) and pi6em2 (c) mouse lines. The presence of both deleted and undeleted PCR products indicate a heterozygous mutant founder that carries just one CRISPR-edited chromosome. d, Genomic sequences of pi17 promoter region in pi17–/– mouse lines. Dashes, genomic sequences deleted by CRISPR; dots, unaltered sequence omitted for clarity.
Extended Data Fig. 2 pi6em1/em1 adult male phenotype.
a, Number of litters produced in 6 months by 2–8 month-old males. b, Body and testis weight of 2–4 month-old pi6em1/em1 and pi6em2/em2 males. Each dot represents an individual mouse. Vertical lines denote median; boxes report 75th and 25th percentiles; whiskers indicate the maximal and minimal values. c, Representative spermatozoa from C57BL/6 and pi6em1/em1 males. d, Representative patterns of meiotic chromosome synapsis in pi6em1/em1 pachytene spermatocytes. SYCP1, Synaptonemal complex protein 1; SYCP3, Synaptonemal complex protein 3. e, Quantification of patterns of meiotic chromosome synapsis depicted in (d) from C57BL/6 (n = 4) and pi6em1/em1 (n = 4) males.
Extended Data Fig. 3 Abundance of transposons in pi6em1/em1 and pi6em2/em2 germ cells.
a, Proportions of the whole genome or piRNA sequences composed of repetitive sequences. b, Abundance of mature piRNAs from the top five major pachytene piRNA-producing loci in indicated cell types measured by small RNA-seq. Each dot represents the abundance of unique-mapping reads in one C57BL/6 (n = 3) or pi6em1/em1 (n = 3) male. Vertical black lines denote median; boxes report 75th and 25th percentiles; whiskers indicate the maximal and minimal values. c, Abundance of transposon-derived RNAs in mouse germ cells. Each dot represents the mean of four (wild-type and pi6em1/em1) or three (pi6em2/em2) biologically independent RNA-seq experiments. Gray dots indicate change in abundance <2-fold and/or FDR > 0.05 determined by DESeq2 (see also Methods).
Extended Data Fig. 4 Pregnancy rate of surrogate mothers in IVF and ICSI experiments.
Percent of pregnant surrogate mothers in IVF (a) and ICSI (b).
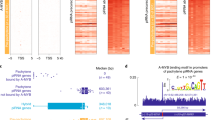
Extended Data Fig. 5 Transcripts directly cleaved by pi6 and pi10-qC2-545.1 piRNAs.
a, Strategy to identify piRNA-directed cleavage sites. b, pi6-dependent cleavage sites in mRNAs or pachytene piRNA precursors from pi10-qC2-545.1 and pi10-qA3-143.1 showing inferred base pairing with the corresponding pi6 piRNA guides. An exemplary piRNA guide is shown where more than one piRNA can direct the same cleavage. c, Cleavage sites in pi6 precursors explained by pi10-qC2-545.1 piRNAs. An exemplary piRNA guide is shown.
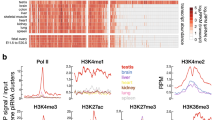
Extended Data Fig. 6 Transcriptome changes in pi6em1/em1 and pi6em2/em2 cells.
a, Expression of mRNAs measured by qRT-PCR using oligo dT(20) to prime cDNA synthesis and PCR primers spanning pi6 piRNA-directed cleavage sites (gene names in red) or designed to detect full-length RNA (gene names in black). Pou2f2 mRNA abundance in spermatids was below the limit of detection by qRT-PCR. b, Abundance of piRNA precursors from the top five major pachytene piRNA-producing loci in indicated cell types measured by RNA-seq. For (a) and (b), thick vertical lines denote median, boxes report 75th and 25th percentiles, and whiskers indicate the maximal and minimal values. Each dot represents an individual mouse.
Supplementary information
Supplementary Information
Supplementary Note
Supplementary Tables
Supplementary Tables 1–6
Supplementary Video 1
C57BL/6 sperm motility at 10 min.
Supplementary Video 3
C57BL/6 sperm motility at 90 min.
Supplementary Video 5
C57BL/6 sperm motility at 3 h.
Supplementary Video 7
C57BL/6 sperm motility at 4 h.
Supplementary Video 9
C57BL/6 sperm motility at 5 h.
Source data
Source Data Fig. 1
Unprocessed gel image.
Rights and permissions
About this article
Cite this article
Wu, PH., Fu, Y., Cecchini, K. et al. The evolutionarily conserved piRNA-producing locus pi6 is required for male mouse fertility. Nat Genet 52, 728–739 (2020). https://doi.org/10.1038/s41588-020-0657-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-020-0657-7
This article is cited by
-
Critical appraisal of the piRNA-PIWI axis in cancer and cancer stem cells
Biomarker Research (2024)
-
piRNA loading triggers MIWI translocation from the intermitochondrial cement to chromatoid body during mouse spermatogenesis
Nature Communications (2024)
-
Jump-starting life: balancing transposable element co-option and genome integrity in the developing mammalian embryo
EMBO Reports (2024)
-
The emerging role of the piRNA/PIWI complex in respiratory tract diseases
Respiratory Research (2023)
-
Genome-wide scan for runs of homozygosity in South American Camelids
BMC Genomics (2023)