Abstract
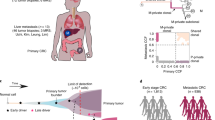
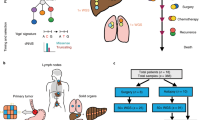
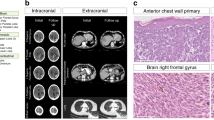
Genetic diversity among metastases is poorly understood but contains important information about disease evolution at secondary sites. Here we investigate inter- and intra-lesion heterogeneity for two types of metastases that associate with different clinical outcomes: lymph node and distant organ metastases in human colorectal cancer. We develop a rigorous mathematical framework for quantifying metastatic phylogenetic diversity. Distant metastases are typically monophyletic and genetically similar to each other. Lymph node metastases, in contrast, display high levels of inter-lesion diversity. We validate these findings by analyzing 317 multi-region biopsies from an independent cohort of 20 patients. We further demonstrate higher levels of intra-lesion heterogeneity in lymph node than in distant metastases. Our results show that fewer primary tumor lineages seed distant metastases than lymph node metastases, indicating that the two sites are subject to different levels of selection. Thus, lymph node and distant metastases develop through fundamentally different evolutionary mechanisms.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Raw polyguanine profiling data and phylogenetic trees for the discovery cohort (Naxerova et al.10) can be downloaded from https://datadryad.org (https://doi.org/10.5061/dryad.vv53d). Original whole-exome sequencing data of Kim et al.11 were deposited to the Sequence Read Archive at the NCBI under the project ID of PRJNA271316. Raw polyguanine profiling data for the new validation cohort are available from https://datadryad.org (https://doi.org/10.5061/dryad.9ghx3ffdf).
Code availability
The source code to calculate the RDS as well as to produce various figure panels is available as jupyter notebooks at http://github.com/johannesreiter/rootdiversity. The notebooks are implemented in Python 3.6. All required input data are contained in Supplementary Tables 1–7.
References
Jones, S. et al. Comparative lesion sequencing provides insights into tumor evolution. Proc. Natl Acad. Sci. USA 105, 4283–4288 (2008).
Mitchell, T. J. et al. Timing the landmark events in the evolution of clear cell renal cell cancer: TRACERx Renal. Cell 173, 611–623.e17 (2018).
Andor, N. et al. Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat. Med. 22, 105–113 (2016).
Cross, W. et al. The evolutionary landscape of colorectal tumorigenesis. Nat. Ecol. Evol. 2, 1661–1672 (2018).
Reiter, J. G. et al. An analysis of genetic heterogeneity in untreated cancers. Nat. Rev. Cancer 19, 639–650 (2019).
Priestley, P. et al. Pan-cancer whole-genome analyses of metastatic solid tumours. Nature 575, 210–216 (2019).
Aceto, N. et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell 158, 1110–1122 (2014).
Yachida, S. et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature 467, 1114–1117 (2010).
Haffner, M. C. et al. Tracking the clonal origin of lethal prostate cancer. J. Clin. Invest. 123, 4918–4922 (2013).
Naxerova, K. et al. Origins of lymphatic and distant metastases in human colorectal cancer. Science 357, 55–60 (2017).
Kim, T.-M. et al. Subclonal genomic architectures of primary and metastatic colorectal cancer based on intratumoral genetic heterogeneity. Clin. Cancer Res. 21, 4461–4472 (2015).
Naxerova, K. et al. Hypermutable DNA chronicles the evolution of human colon cancer. Proc. Natl Acad. Sci. USA 111, E1889–E1898 (2014).
Salipante, S. J. & Horwitz, M. S. Phylogenetic fate mapping. Proc. Natl Acad. Sci. USA 103, 5448–5453 (2006).
Reiter, J. G. et al. Minimal functional driver gene heterogeneity among untreated metastases. Science 361, 1033–1037 (2018).
Shannon, C. E. A mathematical theory of communication. Bell Syst. Tech. J. 27, 379–423 (1948).
Salk, J. J. et al. Clonal expansions in ulcerative colitis identify patients with neoplasia. Proc. Natl Acad. Sci. USA 106, 20871–20876 (2009).
Turajlic, S. et al. Tracking cancer evolution reveals constrained routes to metastases: TRACERx Renal. Cell 173, 581–594.e12 (2018).
Heyde, A., Reiter, J. G., Naxerova, K. & Nowak, M. A. Consecutive seeding and transfer of genetic diversity in metastasis. Proc. Natl Acad. Sci. USA 116, 14129–14137 (2019).
Macintyre, G. et al. How subclonal modeling is changing the metastatic paradigm. Clin. Cancer Res. 23, 630–635 (2017).
Becerra, M. F. et al. Comparative genomic profiling of matched primary and metastatic tumors in renal cell carcinoma. Eur. Urol. Focus 4, 986–994 (2018).
Wagenblast, E. et al. A model of breast cancer heterogeneity reveals vascular mimicry as a driver of metastasis. Nature 520, 358–362 (2015).
Bian, S. et al. Single-cell multiomics sequencing and analyses of human colorectal cancer. Science 362, 1060–1063 (2018).
Ulintz, P. J., Greenson, J. K., Wu, R., Fearon, E. R. & Hardiman, K. M. Lymph node metastases in colon cancer are polyclonal. Clin. Cancer Res. 24, 2214–2224 (2018).
Gundem, G. et al. The evolutionary history of lethal metastatic prostate cancer. Nature 520, 353–357 (2015).
McPherson, A. et al. Divergent modes of clonal spread and intraperitoneal mixing in high-grade serous ovarian cancer. Nat. Genet. 48, 758–767 (2016).
El-Kebir, M., Satas, G. & Raphael, B. J. Inferring parsimonious migration histories for metastatic cancers. Nat. Genet. 50, 718–726 (2018).
Naxerova, K. & Jain, R. K. Using tumour phylogenetics to identify the roots of metastasis in humans. Nat. Rev. Clin. Oncol. 12, 258–272 (2015).
Valastyan, S. & Weinberg, R. A. Tumor metastasis: molecular insights and evolving paradigms. Cell 147, 275–292 (2011).
Ryser, M. D., Min, B.-H., Siegmund, K. D. & Shibata, D. Spatial mutation patterns as markers of early colorectal tumor cell mobility. Proc. Natl Acad. Sci. USA 115, 5774–5779 (2018).
Obenauf, A. C. & Massagué, J. Surviving at a distance: organ specific metastasis. Trends Cancer 1, 76–91 (2015).
Zhao, Z.-M. et al. Early and multiple origins of metastatic lineages within primary tumors. Proc. Natl Acad. Sci. USA 113, 2140–2145 (2016).
Vanharanta, S. & Massagué, J. Origins of metastatic traits. Cancer Cell 24, 410–421 (2013).
Edwards, A. W. F. & Cavalli-Sforza, L. The reconstruction of evolution. Heredity 18, 104–105 (1963).
Felsenstein, J. Inferring Phylogenies (Sinauer Associates, 2003).
Saitou, N. & Nei, M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 (1987).
Paradis, E., Claude, J. & Strimmer, K. APE: Analyses of Phylogenetics and evolution in R language. Bioinformatics 20, 289–290 (2004).
Reiter, J. G. et al. Reconstructing metastatic seeding patterns of human cancers. Nat. Commun. 8, 14114 (2017).
Haeno, H. et al. Computational modeling of pancreatic cancer reveals kinetics of metastasis suggesting optimum treatment strategies. Cell 148, 362–375 (2012).
Durrett, R. Branching Process Models of Cancer (Springer, 2015).
Del Monte, U. Does the cell number 109 still really fit one gram of tumor tissue? Cell Cycle 8, 505–506 (2009).
Acknowledgements
This work was support by grants from the NIH (R37CA225655), AACR (561314) and NHLBI (P01HL142494) to K.N. and from NCI (R00CA22999102) to J.G.R. We thank S. Bian, F. Tang and W. Fu for sharing with us the frequencies of CRC01 subclones across different anatomic sites.
Author information
Authors and Affiliations
Contributions
J.G.R., K.N., W.-T.H., P.G., G.L., I.L., S.D. and E.C.E.W. analyzed data. J.G.R. and S.N. developed the mathematical framework. W.-T.H., P.G. and G.L. performed experiments. W.R.J., M.S.T., A.A.F., H.D.M. and J.K.L. obtained and reviewed clinical samples and clinical data. S.K. and O.K. contributed to data interpretation. K.N. and J.G.R. designed the study. K.N., J.G.R. and W.-T.H. wrote the manuscript with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Distances to tree root (germline).
Mean distances between the root normal sample and samples of primary tumors, lymphatic metastases, and distant metastases, respectively. Distance was measured as the number of internal nodes separating a pair of samples and then normalized by the total number of internal nodes in a given phylogeny. Means are 0.51 for N=16 primary tumors, 0.55 for N=16 lymphatic metastases, and 0.68 for N=16 distant metastases. Box plot elements: center line, median; magenta diamond, mean; box limits, lower and upper quartiles; whiskers, lowest and highest value within 1.5 IQR.
Extended Data Fig. 2 Branch lengths to tree root (germline) for the validation cohort.
Comparison of normalized branch lengths from the normal sample to N=107 primary tumor regions, N=86 lymphatic metastases, and N=34 distant metastases in the validation cohort. Branch lengths were significantly different (p=4.9e-7, Kruskal-Wallis test). Branch lengths for distant metastases were significantly longer than for primary tumor samples (mean 0.9 vs 0.75; p=1.8e-7, Conover’s test) and longer than for lymphatic metastases (mean 0.9 vs 0.77; p=1.9e-6, Conover’s test). Box plot elements: center line, median; magenta diamond, mean; box limits, lower and upper quartiles; whiskers, lowest and highest value within 1.5 IQR.
Extended Data Fig. 3 Spatial classification of primary tumor biopsies.
Spatial classification of primary tumor samples. a, Primary tumor biopsies are classified as luminal or deep by a board-certified pathologist based on established anatomical landmarks. b, Percentages of luminal and deep primary tumor samples in the Naxerova and Reiter/Hung cohorts. c, For each lymphatic and distant metastasis, the closest primary tumor sample is found in the polyguanine marker-based distance matrix. Luminal/deep classifications of closest primary tumor samples are plotted separately for lymphatic and distant metastases. d, As in (c) for the Reiter/Hung cohort. e, as in (c) and (d) for the combined two cohorts. White numbers in panels (b)-(e) denote the number of samples in each group. Two-tailed Fisher’s exact tests were used to calculate the p-values.
Extended Data Fig. 4 Numbers of LN-seeding and DM-seeding clones.
Median RDSLN/RDSDM values for simulations of 10 LN-seeding clones and variable numbers of DM-seeding clones. 100 patients were simulated per parameter combination. The experimentally determined fold change (Naxerova & Reiter/Hung & Kim cohorts) is shown as vertical red line.
Supplementary information
Supplementary Information
Supplementary Figs. 1–32 and Notes
Supplementary Tables
Supplementary Tables 1–7
Rights and permissions
About this article
Cite this article
Reiter, J.G., Hung, WT., Lee, IH. et al. Lymph node metastases develop through a wider evolutionary bottleneck than distant metastases. Nat Genet 52, 692–700 (2020). https://doi.org/10.1038/s41588-020-0633-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-020-0633-2
This article is cited by
-
Multiregion WES of metastatic pancreatic neuroendocrine tumors revealed heterogeneity in genomic alterations, immune microenvironment and evolutionary patterns
Cell Communication and Signaling (2024)
-
Undifferentiated sarcomatoid carcinoma of the pancreas-a single-institution experience with 23 cases
BMC Cancer (2024)
-
Interrogating the roles of lymph node metastasis in systemic immune surveillance
Clinical & Experimental Metastasis (2024)
-
Multiregion sampling of de novo metastatic prostate cancer reveals complex polyclonality and augments clinical genotyping
Nature Cancer (2024)
-
Genomic and evolutionary characteristics of metastatic gastric cancer by routes
British Journal of Cancer (2023)