Abstract
Glaucoma, a disease characterized by progressive optic nerve degeneration, can be prevented through timely diagnosis and treatment. We characterize optic nerve photographs of 67,040 UK Biobank participants and use a multitrait genetic model to identify risk loci for glaucoma. A glaucoma polygenic risk score (PRS) enables effective risk stratification in unselected glaucoma cases and modifies penetrance of the MYOC variant encoding p.Gln368Ter, the most common glaucoma-associated myocilin variant. In the unselected glaucoma population, individuals in the top PRS decile reach an absolute risk for glaucoma 10 years earlier than the bottom decile and are at 15-fold increased risk of developing advanced glaucoma (top 10% versus remaining 90%, odds ratio = 4.20). The PRS predicts glaucoma progression in prospectively monitored, early manifest glaucoma cases (P = 0.004) and surgical intervention in advanced disease (P = 3.6 × 10−6). This glaucoma PRS will facilitate the development of a personalized approach for earlier treatment of high-risk individuals, with less intensive monitoring and treatment being possible for lower-risk groups.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The UKB data are available through the UK Biobank Access Management System https://www.ukbiobank.ac.uk/. The GWAS summary statistics from the glaucoma MTAG analysis is available for research use at https://xikunhan.github.io/site/publication/. We will return the derived data fields following UKB policy; in due course, they will be available through the UK Biobank Access Management System.
References
Weinreb, R. N. & Khaw, P. T. Primary open-angle glaucoma. Lancet 363, 1711–1720 (2004).
Tham, Y.-C. et al. Global prevalence of glaucoma and projections of glaucoma burden through 2040: a systematic review and meta-analysis. Ophthalmology 121, 2081–2090 (2014).
Quigley, H. A. & Broman, A. T. The number of people with glaucoma worldwide in 2010 and 2020. Br. J. Ophthalmol. 90, 262–267 (2006).
Chan, M. P. Y. et al. Glaucoma and intraocular pressure in EPIC-Norfolk Eye Study: cross sectional study. BMJ 358, j3889 (2017).
Mitchell, P., Smith, W., Attebo, K. & Healey, P. R. Prevalence of open-angle glaucoma in Australia. The Blue Mountains Eye Study. Ophthalmology 103, 1661–1669 (1996).
Fraser, S., Bunce, C. & Wormald, R. Risk factors for late presentation in chronic glaucoma. Invest. Ophthalmol. Vis. Sci. 40, 2251–2257 (1999).
Burr, J. M. et al. The clinical effectiveness and cost-effectiveness of screening for open angle glaucoma: a systematic review and economic evaluation. Health Technol. Assess. 11, https://doi.org/10.3310/hta11410 (2007).
Wang, K., Gaitsch, H., Poon, H., Cox, N. J. & Rzhetsky, A. Classification of common human diseases derived from shared genetic and environmental determinants. Nat. Genet. 49, 1319–1325 (2017).
Sanfilippo, P. G., Hewitt, A. W., Hammond, C. J. & Mackey, D. A. The heritability of ocular traits. Surv. Ophthalmol. 55, 561–583 (2010).
Choquet, H. et al. A multiethnic genome-wide association study of primary open-angle glaucoma identifies novel risk loci. Nat. Commun. 9, 2278 (2018).
Leske, M. C., Heijl, A., Hyman, L., Bengtsson, B. & Komaroff, E. Factors for progression and glaucoma treatment: the Early Manifest Glaucoma Trial. Curr. Opin. Ophthalmol. 15, 102–106 (2004).
Garway-Heath, D. F. et al. Latanoprost for open-angle glaucoma (UKGTS): a randomised, multicentre, placebo-controlled trial. Lancet 385, 1295–1304 (2015).
Turley, P. et al. Multi-trait analysis of genome-wide association summary statistics using MTAG. Nat. Genet. 50, 229–237 (2018).
MacGregor, S. et al. Genome-wide association study of intraocular pressure uncovers new pathways to glaucoma. Nat. Genet. 50, 1067–1071 (2018).
Wu, Y., Zheng, Z., Visscher, P. M. & Yang, J. Quantifying the mapping precision of genome-wide association studies using whole-genome sequencing data. Genome Biol. 18, 86 (2017).
Huang, L. et al. Genome-wide analysis identified 17 new loci influencing intraocular pressure in Chinese population. Sci. China Life Sci. 62, 153–164 (2019).
Kuchenbaecker, K. B. et al. Evaluation of polygenic risk scores for breast and ovarian cancer risk prediction in BRCA1 and BRCA2 mutation carriers. J. Natl. Cancer Inst. 109, djw302 (2017).
Lecarpentier, J. et al. Prediction of breast and prostate cancer risks in male BRCA1 and BRCA2 mutation carriers using polygenic risk scores. J. Clin. Oncol. 35, 2240–2250 (2017).
Hewitt, A. W., Mackey, D. A. & Craig, J. E. Myocilin allele-specific glaucoma phenotype database. Hum. Mutat. 29, 207–211 (2008).
Han, X. et al. Myocilin gene Gln368Ter variant penetrance and association with glaucoma in population-based and registry-based studies. JAMA Ophthalmol. 137, 28–35 (2019).
Gharahkhani, P. et al. Accurate imputation-based screening of Gln368Ter myocilin variant in primary open-angle glaucoma. Invest. Ophthalmol. Vis. Sci. 56, 5087–5093 (2015).
Na, J. H. et al. Detection of glaucoma progression by assessment of segmented macular thickness data obtained using spectral domain optical coherence tomography. Invest. Ophthalmol. Vis. Sci. 53, 3817–3826 (2012).
Cheng, C.-Y. et al. Nine loci for ocular axial length identified through genome-wide association studies, including shared loci with refractive error. Am. J. Hum. Genet. 93, 264–277 (2013).
Pickrell, J. K. et al. Detection and interpretation of shared genetic influences on 42 human traits. Nat. Genet. 48, 709–717 (2016).
Verhoeven, V. J. M. et al. Genome-wide meta-analyses of multiancestry cohorts identify multiple new susceptibility loci for refractive error and myopia. Nat. Genet. 45, 314–318 (2013).
Lopes, M. C. et al. Identification of a candidate gene for astigmatism. Invest. Ophthalmol. Vis. Sci. 54, 1260–1267 (2013).
King, R. et al. Genomic locus modulating corneal thickness in the mouse identifies POU6F2 as a potential risk of developing glaucoma. PLoS Genet. 14, e1007145 (2018).
Khawaja, A. P. et al. Genome-wide analyses identify 68 new loci associated with intraocular pressure and improve risk prediction for primary open-angle glaucoma. Nat. Genet. 50, 778–782 (2018).
Gao, X. R., Huang, H., Nannini, D. R., Fan, F. & Kim, H. Genome-wide association analyses identify new loci influencing intraocular pressure. Hum. Mol. Genet. 27, 2205–2213 (2018).
Khera, A. V. et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat. Genet. 50, 1219–1224 (2018).
Dudbridge, F. Power and predictive accuracy of polygenic risk scores. PLoS Genet. 9, e1003348 (2013).
Bycroft, C. et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018).
Bengtsson, B. The variation and covariation of cup and disc diameters. Acta Ophthalmol. (Copenh.) 54, 804–818 (1976).
Han, X. et al. Genome-wide association analysis of 95,549 individuals identifies novel loci and genes influencing optic disc morphology. Hum. Mol. Genet. https://doi.org/10.1093/hmg/ddz193 (2019).
Springelkamp, H. et al. New insights into the genetics of primary open-angle glaucoma based on meta-analyses of intraocular pressure and optic disc characteristics. Hum. Mol. Genet. 26, 438–453 (2017).
Souzeau, E. et al. Australian and New Zealand Registry of Advanced Glaucoma: methodology and recruitment. Clin. Exp. Ophthalmol. 40, 569–575 (2012).
Gharahkhani, P. et al. Common variants near ABCA1, AFAP1 and GMDS confer risk of primary open-angle glaucoma. Nat. Genet. 46, 1120–1125 (2014).
Olsen, C. M. et al. Cohort profile: the QSkin Sun and Health Study. Int. J. Epidemiol. 41, 929–929i (2012).
Wiggs, J. L. et al. The NEIGHBOR consortium primary open-angle glaucoma genome-wide association study: rationale, study design, and clinical variables. J. Glaucoma 22, 517–525 (2013).
Kwon, Y. H., Fingert, J. H., Kuehn, M. H. & Alward, W. L. M. Primary open-angle glaucoma. N. Engl. J. Med. 360, 1113–1124 (2009).
Weinreb, R. N., Garway-Heath, D. F., Leung, C., Medeiros, F. A. & Liebmann, J. Diagnosis of Primary Open Angle Glaucoma: WGA consensus series—10 (Kugler Publications, 2017).
Loh, P.-R. et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat. Genet. 47, 284–290 (2015).
Willer, C. J., Li, Y. & Abecasis, G. R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191 (2010).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Yang, J. et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet. 44, 369–375 (2012).
de Leeuw, C. A., Mooij, J. M., Heskes, T. & Posthuma, D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput. Biol. 11, e1004219 (2015).
Watanabe, K., Taskesen, E., van Bochoven, A. & Posthuma, D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 8, 1826 (2017).
Vilhjálmsson, B. J. et al. Modeling linkage disequilibrium increases accuracy of polygenic risk scores. Am. J. Hum. Genet. 97, 576–592 (2015).
Bulik-Sullivan, B. et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 47, 1236–1241 (2015).
Robin, X. et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 12, 77 (2011).
R Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2017).
Acknowledgements
This work was conducted using the UK Biobank Resource (application no. 25331) and publicly available data from the IGGC. The UK Biobank was established by the Wellcome Trust medical charity, Medical Research Council, Department of Health, Scottish Government and Northwest Regional Development Agency. It also had funding from the Welsh Assembly Government, British Heart Foundation and Diabetes UK. The eye and vision dataset has been developed with additional funding from the National Institute for Health Research Biomedical Research Centre at Moorfields Eye Hospital and the UCL Institute of Ophthalmology, Fight for Sight charity, Moorfields Eye Charity, Macular Society, International Glaucoma Association and Alcon Research Institute. This work was also supported by grants from the National Health and Medical Research Council (NHMRC) of Australia (nos. 1107098, 1116360, 1116495, 1023911, 1150144, 1147571), the Ophthalmic Research Institute of Australia, the BrightFocus Foundation, the UK and Eire Glaucoma Society and charitable funds from the Royal Liverpool University Hospital. S.M., J.E.C., K.P.B., D.A.M. and A.W.H. are supported by NHMRC Fellowships (APP1154543, APP1154824, APP1059954, APP1154513, APP1103329). S.M. was supported by an Australian Research Council Future Fellowship (FT130101902). L.R.P. is supported by National Institutes of Health grant no. R01 EY015473. X.H. is supported by the University of Queensland Research Training Scholarship and Queensland Institute of Medical Research Berghofer PhD Top Up Scholarship. We thank D. Whiteman, R. Neale and C. Olson for providing access to the QSkin samples for use as controls as part of NHMRC grant no. 1063061. We thank S. Wood, J. Pearson and S. Gordon from the Queensland Institute of Medical Research Berghofer Research Institute for their support. The NEIGHBORHOOD consortium is supported by National Institutes of Health grant nos. P30 EY014104, R01 EY015473 and R01 EY022305.
Author information
Authors and Affiliations
Consortia
Contributions
S.M., J.E.C., A.W.H., X.H., P.G., J.L.W. and D.A.M. designed the study and obtained the funding. X.H., A.Q., M.H., J.N.C.B., T.G.K., A.P.K., P.G.H., J.A., H.M., P.G., R.P.I., J.-S.O., T.Z., O.S., M.H.L. and S.M. analyzed the data. J.E.C., X.H., A.Q., M.H., A.P.K., H.M., R.P.I., S.L.G., P.R.H., O.S., E.S., B.S., P.G.H., K.P.B., R.A.M., J.L., J.B.R., A.A., A.G., A.J.R.W., C.E.W., N.A., S.B., A.L.V., I.G., G.R.-S., N.G.M., G.W.M., V.V., R.H., R.W., J.B.J., T.A., L.R.P., A.J.C., S.S., N.A.V., A.C.V., F.P., J.L.H., C.C.W.K., C.M.v.D., R.J.C., P.J.F., P.T.K., C.J.H., D.A.M., P.M., A.J.L., J.L.W., A.W.H. and S.M. contributed to data collection and contributed to genotyping. X.H., J.E.C., A.Q., A.W.H. and S.M. wrote the first draft of the paper. All authors contributed to the final version of the paper.
Corresponding author
Ethics declarations
Competing interests
D.A.M. is consultant/advisor to Allergan, Inc. J.E.C., A.W.H. and S.M. are listed as coinventors on a patent application for the use of genetic risk scores to determine risk and guide treatment.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
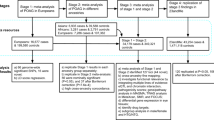
Extended Data Fig. 1 Study design.
We applied the multi-trait analysis of GWAS (MTAG) algorithm to datasets of European descent (unless otherwise specified). a, We applied MTAG to four datasets (glaucoma case-control GWAS from the UKBB; GWAS meta-analysis of intraocular pressure (IOP) from the International Glaucoma Genetics Consortium (IGGC) and the UKBB; Vertical cup-disc ratio (VCDR) GWAS data that was either adjusted for vertical disc diameter (VDD) in the UKBB dataset; or not adjusted for VDD in the IGGC). Novel variants identified through this analysis were then confirmed in two independent data sets: an Australasian cohort of advanced glaucoma (ANZRAG) and a consortium of cohorts from the United States (NEIGHBORHOOD). The clinical significance of the PRS derived from the MTAG analysis was validated in independent samples: first, in advanced glaucoma cases (ANZRAG and samples from Southampton/Liverpool in the UK), and second, in a prospectively monitored clinical cohort with early manifest glaucoma (PROGRESSA). b, Prediction in BMES, where we removed the IGGC VCDR and IGGC IOP GWAS from the training datasets, given that they contain BMES data. c, Prediction in the UKBB glaucoma and ICD-10 POAG cases. Here we removed all glaucoma cases and 3,000 controls with IOP/VCDR measurements as well as their relatives from UKBB VCDR/IOP GWAS. We also evaluated the performance of PRS in non-European ancestry (192 cases and 6,841 controls of South Asian ancestry in UKBB). d, Cumulative risk of glaucoma in UKBB. For the analysis of MYOC p.Gln368Ter carriers (n = 965; cases = 72; controls = 893), participants were stratified into tertiles of PRS. We also examined cumulative risk of glaucoma in the general population (that is in MYOC p.Gln368Ter non-carriers, n = 381,196; cases = 7,381; controls = 373,815) stratifying by deciles of the PRS. The discovery and testing datasets were designed to derive the PRS with no sample overlap (Supplementary Note).
Supplementary information
Supplementary Information
Supplementary Note, Figs. 1–13 and Tables 1–13
Rights and permissions
About this article
Cite this article
Craig, J.E., Han, X., Qassim, A. et al. Multitrait analysis of glaucoma identifies new risk loci and enables polygenic prediction of disease susceptibility and progression. Nat Genet 52, 160–166 (2020). https://doi.org/10.1038/s41588-019-0556-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-019-0556-y
This article is cited by
-
Automated vertical cup-to-disc ratio determination from fundus images for glaucoma detection
Scientific Reports (2024)
-
Polygenic risk score-based phenome-wide association for glaucoma and its impact on disease susceptibility in two large biobanks
Journal of Translational Medicine (2024)
-
Cis-meQTL for cocaine use-associated DNA methylation in an HIV-positive cohort show pleiotropic effects on multiple traits
BMC Genomics (2023)
-
Genetic Risk Assessment of Degenerative Eye Disease (GRADE): study protocol of a prospective assessment of polygenic risk scores to predict diagnosis of glaucoma and age-related macular degeneration
BMC Ophthalmology (2023)
-
Identification of the potential association between SARS-CoV-2 infection and acute kidney injury based on the shared gene signatures and regulatory network
BMC Infectious Diseases (2023)