Abstract
The poor outcomes in esophageal adenocarcinoma (EAC) prompted us to interrogate the pattern and timing of metastatic spread. Whole-genome sequencing and phylogenetic analysis of 388 samples across 18 individuals with EAC showed, in 90% of patients, that multiple subclones from the primary tumor spread very rapidly from the primary site to form multiple metastases, including lymph nodes and distant tissues—a mode of dissemination that we term ‘clonal diaspora’. Metastatic subclones at autopsy were present in tissue and blood samples from earlier time points. These findings have implications for our understanding and clinical evaluation of EAC.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Sequencing data that support the findings of this paper have been deposited in the European Genome-phenome Archive with the accession code EGAD00001005434.
Code availability
All code required to reproduce the analysis outlined in this manuscript can be found in the Methods and Supplementary Methods. There are no restrictions to the accessibility of this code.
References
Sporn, M. B. The war on cancer. Lancet 347, 1377–1381 (1996).
Waterman, T. A. et al. The prognostic importance of immunohistochemically detected node metastases in resected esophageal adenocarcinoma. Ann. Thorac. Surg. 78, 1161–1169 (2004).
Matsuda, S., Takeuchi, H., Kawakubo, H. & Kitagawa, Y. Three-field lymph node dissection in esophageal cancer surgery. J. Thorac. Dis. 9, S731–S740 (2017).
Lou, F. et al. Esophageal cancer recurrence patterns and implications for surveillance. J. Thorac. Oncol. 8, 1558–1562 (2013).
Smyth, E. C. et al. Oesophageal cancer. Nat. Rev. Dis. Primers 3, 17048 (2017).
Cunningham, D. et al. Capecitabine and oxaliplatin for advanced esophagogastric cancer. N. Engl. J. Med. 358, 36–46 (2008).
Greaves, M. & Maley, C. C. Clonal evolution in cancer. Nature 481, 306–313 (2012).
Klein, C. A. Parallel progression of primary tumours and metastases. Nat. Rev. Cancer 9, 302–312 (2009).
Davis, A., Gao, R. & Navin, N. Tumor evolution: linear, branching, neutral or punctuated? Biochim. Biophys. Acta 1867, 151–161 (2017).
Yates, L. R. & Campbell, P. J. Evolution of the cancer genome. Nat. Rev. Genet. 13, 795–806 (2012).
Nik-Zainal, S. et al. The life history of 21 breast cancers. Cell 149, 994–1007 (2012).
Murugaesu, N. et al. Tracking the genomic evolution of esophageal adenocarcinoma through neoadjuvant chemotherapy. Cancer Discov. 5, 821–831 (2015).
Gerstung, M. et al. The evolutionary history of 2,658 cancers. Preprint at bioRxiv https://doi.org/10.1101/161562 (2018).
Secrier, M. et al. Mutational signatures in esophageal adenocarcinoma define etiologically distinct subgroups with therapeutic relevance. Nat. Genet. 48, 1131–1141 (2016).
Dulak, A. M. et al. Gastrointestinal adenocarcinomas of the esophagus, stomach, and colon exhibit distinct patterns of genome instability and oncogenesis. Cancer Res. 72, 4383–4393 (2012).
Weaver, J. M. et al. Ordering of mutations in preinvasive disease stages of esophageal carcinogenesis. Nat. Genet. 46, 837–843 (2014).
Ross-Innes, C. S. et al. Whole-genome sequencing provides new insights into the clonal architecture of Barrett’s esophagus and esophageal adenocarcinoma. Nat. Genet. 47, 1038–1046 (2015).
Dulak, A. M. et al. Exome and whole-genome sequencing of esophageal adenocarcinoma identifies recurrent driver events and mutational complexity. Nat. Genet. 45, 478–486 (2013).
Nones, K. et al. Genomic catastrophes frequently arise in esophageal adenocarcinoma and drive tumorigenesis. Nat. Commun. 5, 5224 (2014).
Frankell, A. M. et al. The landscape of selection in 551 esophageal adenocarcinomas defines genomic biomarkers for the clinic. Nat. Genet. 51, 506–516 (2019).
Yates, L. R. et al. Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nat. Med. 21, 751–759 (2015).
Rodriguez-Martin, B. et al. Pan-cancer analysis of whole genomes reveals driver rearrangements promoted by LINE-1 retrotransposition in human tumours. Preprint at bioRxiv https://doi.org/10.1101/179705 (2018).
Ajani, J. A. et al. Esophageal and esophagogastric junction cancers, version 1.2015. J. Natl Compr. Canc. Netw. 13, 194–227 (2015).
Yachida, S. et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature 467, 1114–1117 (2010).
Sottoriva, A. et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc. Natl Acad. Sci. USA 110, 4009–4014 (2013).
Mariette, C. et al. Pattern of recurrence following complete resection of esophageal carcinoma and factors predictive of recurrent disease. Cancer 97, 1616–1623 (2003).
Alexandrov, L. B. et al. Signatures of mutational processes in human cancer. Nature 500, 415–421 (2013).
Liu, D. et al. Mutational patterns in chemotherapy resistant muscle-invasive bladder cancer. Nat. Commun. 8, 2193 (2017).
Behjati, S. et al. Mutational signatures of ionizing radiation in second malignancies. Nat. Commun. 7, 12605 (2016).
Dentro, S. C. et al. Portraits of genetic intra-tumour heterogeneity and subclonal selection across cancer types. Preprint at bioRxiv https://doi.org/10.1101/312041 (2018).
Lodato, M. A. et al. Aging and neurodegeneration are associated with increased mutations in single human neurons. Science 359, 555–559 (2018).
Gao, Z., Wyman, M. J., Sella, G. & Przeworski, M. Interpreting the dependence of mutation rates on age and time. PLoS Biol. 14, e1002355 (2016).
Letouze, E. et al. Mutational signatures reveal the dynamic interplay of risk factors and cellular processes during liver tumorigenesis. Nat. Commun. 8, 1315 (2017).
Blokzijl, F. et al. Tissue-specific mutation accumulation in human adult stem cells during life. Nature 538, 260–264 (2016).
Alexandrov, L. B. et al. Clock-like mutational processes in human somatic cells. Nat. Genet. 47, 1402–1407 (2015).
Pienta, K. J., Robertson, B. A., Coffey, D. S. & Taichman, R. S. The cancer diaspora: metastasis beyond the seed and soil hypothesis. Clin. Cancer Res. 19, 5849–5855 (2013).
Gundem, G. et al. The evolutionary history of lethal metastatic prostate cancer. Nature 520, 353–357 (2015).
Foulds, L. The experimental study of tumor progression: a review. Cancer Res. 14, 327–339 (1954).
Naxerova, K. et al. Origins of lymphatic and distant metastases in human colorectal cancer. Science 357, 55–60 (2017).
Sottoriva, A. et al. A Big Bang model of human colorectal tumor growth. Nat. Genet. 47, 209–216 (2015).
Sjoquist, K. M. et al. Survival after neoadjuvant chemotherapy or chemoradiotherapy for resectable oesophageal carcinoma: an updated meta-analysis. Lancet Oncol. 12, 681–692 (2011).
Gabriel, E. et al. Novel calculator to estimate overall survival benefit from neoadjuvant chemoradiation in patients with esophageal adenocarcinoma. J. Am. Coll. Surg. 224, 884–894.e1 (2017).
Burt, B. M. et al. Utility of adjuvant chemotherapy after neoadjuvant chemoradiation and esophagectomy for esophageal cancer. Ann. Surg. 266, 297–304 (2017).
Pasquali, S. et al. Survival after neoadjuvant and adjuvant treatments compared to surgery alone for resectable esophageal carcinoma: a network meta-analysis. Ann. Surg. 265, 481–491 (2017).
Parikh, A. R. et al. Liquid versus tissue biopsy for detecting acquired resistance and tumor heterogeneity in gastrointestinal cancers. Nat. Med. 25, 1415–1421 (2019).
Van Roy, N. et al. Shallow whole genome sequencing on circulating cell-free DNA allows reliable noninvasive copy-number profiling in neuroblastoma patients. Clin. Cancer Res. 23, 6305–6314 (2017).
Hu, Z. et al. Quantitative evidence for early metastatic seeding in colorectal cancer. Nat. Genet. 51, 1113–1122 (2019).
Robinson, D. R. et al. Integrative clinical genomics of metastatic cancer. Nature 548, 297–303 (2017).
Bolli, N. et al. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nat. Commun. 5, 2997 (2014).
Martincorena, I. et al. Universal patterns of selection in cancer and somatic tissues. Cell 171, 1029–1041.e21 (2017).
Japanese Gastric Cancer Association. Japanese classification of gastric carcinoma: 3rd English edition. Gastric Cancer 14, 101–112 (2011).
Jiao, W., Vembu, S., Deshwar, A. G., Stein, L. & Morris, Q. Inferring clonal evolution of tumors from single nucleotide somatic mutations. BMC Bioinformatics 15, 35 (2014).
Kuipers, J., Jahn, K., Raphael, B. J. & Beerenwinkel, N. Single-cell sequencing data reveal widespread recurrence and loss of mutational hits in the life histories of tumors. Genome Res. 27, 1885–1894 (2017).
Acknowledgements
Above all, we are indebted to the patients who donated tissue samples to this project and thank them and their families who supported them through it. We also thank the following individuals for help with study set-up, patient liaison and tissue collection: B. Smith, N. Chinyama, V. Sujendran, P. Safranek, A. Xanthos, T. Nuckcheddy-Grant, R. de la Rue, S. Zeki, R. Fels Elliott, P. Collins, K. Puttock, S. Rabey and staff at Arthur Rank Hospice and L. A. Wylie for scientific discussion and contribution. We thank the OCCAMS Consortium for providing the vehicle through which funding for the ICGC was obtained. We are grateful to S. Tavaré (a Fellow of the Royal Society) for guidance and support for the esophageal WGS project as a part of the ICGC. We also thank J. Westmoreland (LMB Visual Aids) for graphic art expertise. Thanks also go to the Cancer Research UK Cambridge Institute Genomics Core for technical expertise. We thank the Human Research Tissue Bank, which is supported by the National Institute for Health Research (NIHR) Cambridge Biomedical Research Centre, at Addenbrooke’s Hospital. Additional infrastructure support was provided by the Cancer Research UK-funded Experimental Cancer Medicine Centre in Cambridge. Computation by D.C.W. used the Oxford Biomedical Research Computing facility—a joint development between the Wellcome Centre for Human Genetics and the Big Data Institute, supported by Health Data Research UK and the NIHR Oxford Biomedical Research Centre. A.N. was funded through an MRC Clinical Research Fellowship. The work (including 1× WGS sequencing) was funded through the above, an MRC core grant (RG84369) and an NIHR Research Professorship (RG67258) to R.C.F. Funding for sample sequencing (50× WGS) was through the ICGC, and was funded by a program grant from Cancer Research UK (RG66287). All OCCAMS samples that were part of the surgical/endoscopy cohort were obtained from Cambridge patients. D.C.W. is funded by the Li Ka Shing Foundation and NIHR Oxford Biomedical Research Centre.
Author information
Authors and Affiliations
Contributions
A.N. designed and implemented the rapid autopsy study, collected the samples, performed the experiments, analyzed the data and wrote the manuscript. M.G. and S.D.P. contributed expertise in pathology and sample collection for the rapid autopsy study. I.D.-B. and N.G. assisted in study implementation and, along with J.C., assisted with sample collection at autopsy. M.S. performed the structural variant analysis. M.D.E. performed genomic data generation and quality control. L.B. conducted data management. X.L., P.L.-S. and J.W. were involved with autopsy sample collection, advice on experiments and data analysis. X.L. contributed to the experiments, paper writing and figure design. L.B.A. and I.M. assisted with data analysis. N.G. assisted with study implementation. S. MacRae coordinated the sequencing of samples from the OCCAMS project and contributed to paper writing. S. Malhotra and A.M. provided pathology data. T.T., S.G., L.P. and D.G. assisted in implementation and ethical conduct of the autopsy study. R.H.H. and A.H. were involved in surgical sample collection and providing surgical expertise. M.R.S. contributed to critical evaluation of the study data and manuscript. D.C.W. was responsible for data analysis, paper writing and assuring integrity of the data. The OCCAMS Consortium was the vehicle through which the infrastructure and funding was obtained to support the study, and this consortium contributed to discussions on the ICGC data and the clinical ramifications. R.C.F. provided grant funding and was responsible for designing the study, supervising the project, writing the paper and assuring integrity of the data.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Flowchart describing key steps taken to construct phylogenetic trees.
A. The details of phylogenetic tree reconstruction is further elaborated in Supplementary methods, Mutation clustering and phylogenetic tree construction (p.25).
Extended Data Fig. 2 Phylogenetic tree construction for example case S3.
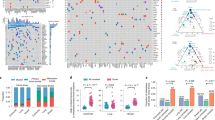
1) Battenberg algorithm to determine total copy number (purple line) and minor allele (blue line). Y-axis =number of chromosome copies, X-axis= chromosome and position. The average ploidy, aberrant cell fraction (cellularity) and goodness of fit to the model are shown for each sample, Primary E1, E2, Lymph node L1 and Distant metastasis D1. The goodness of fit is a measure of the amount of the genome with clonal, rather than subclonal copy number states. D1 has a subclonal mix of different copy number states resulting in noninteger total copy number, for example on chromosome 2, resulting in a goodness of fit below 100%. 2) Bayesian Dirichlet Process to cluster SNVs based on CCF in each sample. The density plots show the posterior probability of a mutational cluster, these are produced for every pair of samples and selected plots are shown High density at CCF of (0,0) indicates subclones that are not present in the pair of samples shown in a particular plot. 3) Clustering of results—Clusters are identified as local maxima in the posterior density. The table shows the number of SNVs assigned to each cluster, and their associated CCFs. 4) Unscaled Tree construction using the sum rule and crossing rule as detailed in Supplementary Methods p25. 5) Final Tree -The tree is drawn as seen in Fig. 2 and Extended Data2, branch lengths are proportional to the number of SNVs assigned to each subclone. Scales vary on a per case basis depending on the total number of SNVs, in order to fit cases on one figure. Trees are annotated with the gene names of known drivers, and the colour of each branch represents a trunk (pink), branch (purple) or leaf (yellow). The grey circles represent clones and subclones and their CCFs are shown in Supplementary Table 5 and 6.
Extended Data Fig. 3 Phylogenetic trees of cases in cohort with only nodal or distant organ disease, as derived from H-WGS.
E=esophagus, D=distant organ, L=lymph node, B= Barrett’s. For precise anatomical locations, refer to Supplementary Tables 3 and 4. MRCA=most recent common ancestor. Pink=trunk (shared events), Purple=branch (shared by more than one sample), Yellow=leaf (unique to one sample). Grey dots at the end of the lines (truncal, branches or leaves) represent subclones or clones, whose CCFs are shown in Supplementary Table 5 and 6. Trees are annotated with key driver events as identified from the literature14,16,19. Black=point mutations, Red=copy number alterations, purple= structural variants. The adjacent scales are relative to the number of SNVs in that particular case and hence constructed on a case by case basis.
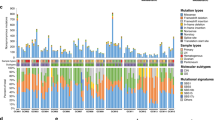
Extended Data Fig. 4 Structural variation of 18 metastatic esophageal adenocarcinoma cases.
a. Similarity matrix based clustering for all SVs 122 genomes across 19 cases. SVs were deemed to refer to the same rearrangement event across cases if their corresponding breakpoint locations fell within a window of maximum 50 bp. The individual sample types are shown as a separate row on the x axis with the color key depicting the sample type. The purple scale indicates the number of shared SVs. (L=lymph nodes; M=metastasis; T=tumor). b. Histogram showing the percentage of rearranged genes that are concordant, unique to tumors and unique to metastases. Two-tailed Welch test P=0.2674 demonstrating no overall difference between total number of SVs in primary, local lymph nodes and distant metastases c. Stacked bar charts showing the composition of various SVs in each sample on a per patient basis INV= inversion, ME= mobile element, BND= translocation DEL=deletion, DUP=duplication, INS= insertion.
Extended Data Fig. 5 Random simulation model for 1xWGS cluster detection.
The number of mutations detected correlates strongly with the CCF of the cluster (Pearson r=0.992, n=100). Number of mutations in each cluster =1000.
Extended Data Fig. 6 Correlation of fraction of mutations detected with CCF as a function of cluster size using simulated 1xWGS data.
Pearson correlation coefficient is above 0.97 for clusters with 200 or more mutations.
Extended Data Fig. 7
Bar chart demonstrating the Pearson correlation coefficient of VAF at 1xWGS and High Depth Resequencing (n=33).
Extended Data Fig. 8 Detection of Selection in subsets of mutations.
SNVs and indels from all cases (n=18) were aggregated into 4 different subsets: clonal = variants found in the MRCA (n=378453); subclonal = variants not found in the MRCA (n=516136); pre-diaspora = variants found above the diaspora founder clone in the phylogenetic tree (n=313545); post-diaspora = variants found in the diaspora founder or in clones below the founder in the phylogenetic tree (n=295316). Within each subset, dN/dS analysis was performed separately on: missense variants; truncating variants. Bars show maximum likelihood estimates of dN/dS values, with values greater than 1 (dashed line) indicating positive selection. Vertical lines = 95% confidence intervals, estimated using Wald test.
Extended Data Fig. 9 Percentage of truncal and branch clusters in tissue from earlier time-points.
Stacked horizontal bar chart representing the percentage of truncal and branch clusters present in tissue from earlier time-points on the x-axis and the Case ID on the y-axis. P1 diagnosis* is a frozen sample, while the rest are FFPE. Blue = truncal, maroon = branch, grey = not present. The number of clusters (n) is demonstrated for each case.
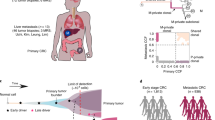
Extended Data Fig. 10 ctDNA analysis from historical plasma samples.
Digital PCR traces of mutant allele fraction for TP53 on the Y-axis and days from diagnosis on the X-axis, and grey areas indicate periods of therapy. Where subclones and clones are seen at 1xWGS on plasma, they are highlighted on the 50x phylogenetic tree (coloured blue). The samples in which these subcloens and clones are present in are shown in Supplementary Table 3. There was no TP53 data for S3 as it was wild type for TP53 mutations. Copy number traces for P1 are shown, with the arrow demonstrating an area of MET amplification.
Supplementary information
Supplementary Information
Supplementary Figs. 1–5, Tables 1–4, 10 and 12–15, and Supplementary Note
Supplementary Tables
Supplementary Tables 5–9, 11 and 16
Rights and permissions
About this article
Cite this article
Noorani, A., Li, X., Goddard, M. et al. Genomic evidence supports a clonal diaspora model for metastases of esophageal adenocarcinoma. Nat Genet 52, 74–83 (2020). https://doi.org/10.1038/s41588-019-0551-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-019-0551-3
This article is cited by
-
How much do we know about the metastatic process?
Clinical & Experimental Metastasis (2024)
-
Spatial biology of cancer evolution
Nature Reviews Genetics (2023)
-
Genomic and evolutionary characteristics of metastatic gastric cancer by routes
British Journal of Cancer (2023)
-
Evolutionary route of nasopharyngeal carcinoma metastasis and its clinical significance
Nature Communications (2023)
-
Metabolic tumour and nodal response to neoadjuvant chemotherapy on FDG PET-CT as a predictor of pathological response and survival in patients with oesophageal adenocarcinoma
European Radiology (2023)