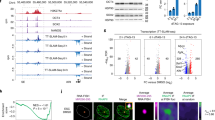
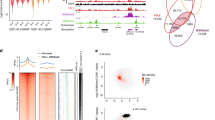
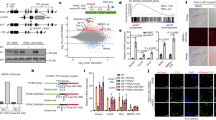
Abstract
Chromatin architecture has been implicated in cell type-specific gene regulatory programs, yet how chromatin remodels during development remains to be fully elucidated. Here, by interrogating chromatin reorganization during human pluripotent stem cell (hPSC) differentiation, we discover a role for the primate-specific endogenous retrotransposon human endogenous retrovirus subfamily H (HERV-H) in creating topologically associating domains (TADs) in hPSCs. Deleting these HERV-H elements eliminates their corresponding TAD boundaries and reduces the transcription of upstream genes, while de novo insertion of HERV-H elements can introduce new TAD boundaries. The ability of HERV-H to create TAD boundaries depends on high transcription, as transcriptional repression of HERV-H elements prevents the formation of boundaries. This ability is not limited to hPSCs, as these actively transcribed HERV-H elements and their corresponding TAD boundaries also appear in pluripotent stem cells from other hominids but not in more distantly related species lacking HERV-H elements. Overall, our results provide direct evidence for retrotransposons in actively shaping cell type- and species-specific chromatin architecture.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All sequencing datasets have been deposited in the Gene Expression Omnibus repository with the accession number GSE116862.
Code availability
Scripts are available at https://github.com/shawnzhangyx/cvdc_scripts.
References
Dekker, J. & Mirny, L. The 3D genome as moderator of chromosomal communication. Cell 164, 1110–1121 (2016).
Yu, M. & Ren, B. The three-dimensional organization of mammalian genomes. Annu. Rev. Cell Dev. Biol. 33, 265–289 (2017).
Meaburn, K. J. & Misteli, T. Cell biology: chromosome territories. Nature 445, 379–781 (2007).
Lieberman-Aiden, E. et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 (2009).
Dixon, J. R. et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012).
Sexton, T. et al. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 148, 458–472 (2012).
Nora, E. P. et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485, 381–385 (2012).
Dixon, J. R. et al. Chromatin architecture reorganization during stem cell differentiation. Nature 518, 331–336 (2015).
Dixon, J. R., Gorkin, D. U. & Ren, B. Chromatin domains: the unit of chromosome organization. Mol. Cell 62, 668–680 (2016).
Rao, S. S. et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014).
Jin, F. et al. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature 503, 290–294 (2013).
Lupianez, D. G. et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene–enhancer interactions. Cell 161, 1012–1025 (2015).
Lupianez, D. G., Spielmann, M. & Mundlos, S. Breaking TADs: how alterations of chromatin domains result in disease. Trends Genet. 32, 225–237 (2016).
Ibn-Salem, J. et al. Deletions of chromosomal regulatory boundaries are associated with congenital disease. Genome Biol. 15, 423 (2014).
Franke, M. et al. Formation of new chromatin domains determines pathogenicity of genomic duplications. Nature 538, 265–269 (2016).
Nasmyth, K. Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu Rev. Genet. 35, 673–745 (2001).
Alipour, E. & Marko, J. F. Self-organization of domain structures by DNA-loop-extruding enzymes. Nucleic Acids Res. 40, 11202–11212 (2012).
Rao, S. S. P. et al. Cohesin loss eliminates all loop domains. Cell 171, 305–320.e24 (2017).
Pombo, A. & Dillon, N. Three-dimensional genome architecture: players and mechanisms. Nat. Rev. Mol. Cell Biol. 16, 245–257 (2015).
Nora, E. P. et al. Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell 169, 930–944.e22 (2017).
Vietri Rudan, M. et al. Comparative Hi-C reveals that CTCF underlies evolution of chromosomal domain architecture. Cell Rep. 10, 1297–1309 (2015).
Schmidt, D. et al. Waves of retrotransposon expansion remodel genome organization and CTCF binding in multiple mammalian lineages. Cell 148, 335–348 (2012).
Bonev, B. et al. Multiscale 3D genome rewiring during mouse neural development. Cell 171, 557–572 (2017).
Stadhouders, R. et al. Transcription factors orchestrate dynamic interplay between genome topology and gene regulation during cell reprogramming. Nat. Genet. 50, 238–249 (2018).
Krijger, P. H. et al. Cell-of-origin-specific 3D genome structure acquired during somatic cell reprogramming. Cell Stem Cell 18, 597–610 (2016).
Beagan, J. A. et al. Local genome topology can exhibit an incompletely rewired 3D-folding state during somatic cell reprogramming. Cell Stem Cell 18, 611–624 (2016).
Veevers, J. et al. Cell-surface marker signature for enrichment of ventricular cardiomyocytes derived from human embryonic stem cells. Stem Cell Rep. 11, 828–841 (2018).
Fields, P. A. et al. Dynamic reorganization of nuclear architecture during human cardiogenesis. Preprint at bioRxiv https://www.biorxiv.org/content/10.1101/222877v1 (2017).
Crane, E. et al. Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature 523, 240–244 (2015).
Grow, E. J. et al. Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells. Nature 522, 221–225 (2015).
Goke, J. et al. Dynamic transcription of distinct classes of endogenous retroviral elements marks specific populations of early human embryonic cells. Cell Stem Cell 16, 135–141 (2015).
Lu, X. et al. The retrovirus HERVH is a long noncoding RNA required for human embryonic stem cell identity. Nat. Struct. Mol. Biol. 21, 423–425 (2014).
Romer, C., Singh, M., Hurst, L. D. & Izsvak, Z. How to tame an endogenous retrovirus: HERVH and the evolution of human pluripotency. Curr. Opin. Virol. 25, 49–58 (2017).
Santoni, F. A., Guerra, J. & Luban, J. HERV-H RNA is abundant in human embryonic stem cells and a precise marker for pluripotency. Retrovirology 9, 111 (2012).
Ohnuki, M. et al. Dynamic regulation of human endogenous retroviruses mediates factor-induced reprogramming and differentiation potential. Proc. Natl Acad. Sci. USA 111, 12426–12431 (2014).
Loewer, S. et al. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat. Genet. 42, 1113–1117 (2010).
Xie, W. et al. Epigenomic analysis of multilineage differentiation of human embryonic stem cells. Cell 153, 1134–1148 (2013).
Greenwald, W. W. et al. Subtle changes in chromatin loop contact propensity are associated with differential gene regulation and expression. Nat. Commun. 10, 1054 (2019).
Lengronne, A. et al. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature 430, 573–578 (2004).
Busslinger, G. A. et al. Cohesin is positioned in mammalian genomes by transcription, CTCF and Wapl. Nature 544, 503–507 (2017).
Jacques, P. E., Jeyakani, J. & Bourque, G. The majority of primate-specific regulatory sequences are derived from transposable elements. PLoS Genet. 9, e1003504 (2013).
Barakat, T. S. et al. Functional dissection of the enhancer repertoire in human embryonic stem cells. Cell Stem Cell 23, 276–288.e8 (2018).
Wang, J. et al. Primate-specific endogenous retrovirus-driven transcription defines naive-like stem cells. Nature 516, 405–409 (2014).
Ng, S. Y., Johnson, R. & Stanton, L. W. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 31, 522–533 (2012).
Liu, J., Li, Y., Lin, B., Sheng, Y. & Yang, L. HBL1 is a human long noncoding RNA that modulates cardiomyocyte development from pluripotent stem cells by counteracting MIR1. Dev. Cell 42, 333–348.e5 (2017).
Koyanagi-Aoi, M. et al. Differentiation-defective phenotypes revealed by large-scale analyses of human pluripotent stem cells. Proc. Natl Acad. Sci. USA 110, 20569–20574 (2013).
Goodchild, N. L., Wilkinson, D. A. & Mager, D. L. Recent evolutionary expansion of a subfamily of RTVL-H human endogenous retrovirus-like elements. Virology 196, 778–788 (1993).
Mager, D. L. & Freeman, J. D. HERV-H endogenous retroviruses: presence in the New World branch but amplification in the Old World primate lineage. Virology 213, 395–404 (1995).
Kijima, T. E. & Innan, H. On the estimation of the insertion time of LTR retrotransposable elements. Mol. Biol. Evol. 27, 896–904 (2010).
Britten, R. J. & Davidson, E. H. Repetitive and non-repetitive DNA sequences and a speculation on the origins of evolutionary novelty. Q. Rev. Biol. 46, 111–138 (1971).
King, M. C. & Wilson, A. C. Evolution at two levels in humans and chimpanzees. Science 188, 107–116 (1975).
Chuong, E. B., Elde, N. C. & Feschotte, C. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science 351, 1083–1087 (2016).
Wang, Y. et al. Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev. Cell 25, 69–80 (2013).
De Cecco, M. et al. L1 drives IFN in senescent cells and promotes age-associated inflammation. Nature 566, 73–78 (2019).
Kong, Y. et al. Transposable element expression in tumors is associated with immune infiltration and increased antigenicity. Preprint at bioRxiv https://www.biorxiv.org/content/10.1101/388215v1 (2018).
Reilly, M. T., Faulkner, G. J., Dubnau, J., Ponomarev, I. & Gage, F. H. The role of transposable elements in health and diseases of the central nervous system. J. Neurosci. 33, 17577–17586 (2013).
Lian, X. et al. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/β-catenin signaling under fully defined conditions. Nat. Protoc. 8, 162–175 (2013).
Hashem, S. I. et al. Brief Report: oxidative stress mediates cardiomyocyte apoptosis in a human model of Danon disease and heart failure. Stem Cells 33, 2343–2350 (2015).
Shen, Y. et al. A map of the cis-regulatory sequences in the mouse genome. Nature 488, 116–120 (2012).
Gorkin, D. et al. Systematic mapping of chromatin state landscapes during mouse development. Preprint at bioRxiv https://www.biorxiv.org/content/10.1101/166652v1 (2017).
Jolma, A. et al. DNA-binding specificities of human transcription factors. Cell 152, 327–339 (2013).
Yan, J. et al. Transcription factor binding in human cells occurs in dense clusters formed around cohesin anchor sites. Cell 154, 801–813 (2013).
Liu, Z. et al. Efficient CRISPR/Cas9-mediated versatile, predictable, and donor-free gene knockout in human pluripotent stem cells. Stem Cell Rep. 7, 496–507 (2016).
Haeussler, M. et al. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 17, 148 (2016).
Kim, S., Kim, D., Cho, S. W., Kim, J. & Kim, J. S. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 24, 1012–1019 (2014).
Zuris, J. A. et al. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat. Biotechnol. 33, 73–80 (2015).
Santos, D. P., Kiskinis, E., Eggan, K. & Merkle, F. T. Comprehensive protocols for CRISPR/Cas9-based gene editing in human pluripotent stem cells. Curr. Protoc. Stem Cell Biol. 38, 5B.6.1–5B.6.60 (2016).
Radzisheuskaya, A., Shlyueva, D., Muller, I. & Helin, K. Optimizing sgRNA position markedly improves the efficiency of CRISPR/dCas9-mediated transcriptional repression. Nucleic Acids Res. 44, e141 (2016).
Konermann, S. et al. Genome-scale transcriptional activation by an engineered CRISPR–Cas9 complex. Nature 517, 583–588 (2015).
Xie, S., Duan, J., Li, B., Zhou, P. & Hon, G. C. Multiplexed engineering and analysis of combinatorial enhancer activity in single cells. Mol. Cell 66, 285–299.e5 (2017).
Ding, S. et al. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell 122, 473–483 (2005).
Meir, Y. J. et al. Genome-wide target profiling of piggyBac and Tol2 in HEK 293: pros and cons for gene discovery and gene therapy. BMC Biotechnol. 11, 28 (2011).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Ramirez, F., Dundar, F., Diehl, S., Gruning, B. A. & Manke, T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 42, W187–W191 (2014).
Dobin, A. et al. STAR: ultrafast universal RNA-Seq aligner. Bioinformatics 29, 15–21 (2013).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Durand, N. C. et al. Juicebox provides a visualization system for Hi-C contact maps with unlimited zoom. Cell Syst. 3, 99–101 (2016).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Casper, J. et al. The UCSC Genome Browser database: 2018 update. Nucleic Acids Res. 46, D762–D769 (2018).
Xu, C. & Corces, V. G. Nascent DNA methylome mapping reveals inheritance of hemimethylation at CTCF/cohesin sites. Science 359, 1166–1170 (2018).
DePristo, M. A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498 (2011).
Selvaraj, S., R Dixon, J., Bansal, V. & Ren, B. Whole-genome haplotype reconstruction using proximity-ligation and shotgun sequencing. Nat. Biotechnol. 31, 1111–1118 (2013).
Edge, P., Bafna, V. & Bansal, V. HapCUT2: robust and accurate haplotype assembly for diverse sequencing technologies. Genome Res. 27, 801–812 (2017).
Greenwald, W. W. et al. Pgltools: a genomic arithmetic tool suite for manipulation of Hi-C peak and other chromatin interaction data. BMC Bioinformatics 18, 207 (2017).
Hu, T. T. et al. The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat. Genet. 43, 476–481 (2011).
Slotte, T. et al. The Capsella rubella genome and the genomic consequences of rapid mating system evolution. Nat. Genet. 45, 831–835 (2013).
Acknowledgements
We thank S. Kuan and B. Li for sequencing and bioinformatic support. We thank E. Nostrand for RNA extraction. We thank M. Daadi (University of Texas Health Science Center at San Antonio) for providing marmoset iPSCs. We thank F. Gage (Salk Institute) for providing chimpanzee and bonobo iPSCs. This project is supported by funding from the Ludwig Institute for Cancer Research (to B.R.) and NIH (1UM1HL128773 to S.M.E., N.C.C., E.D. and B.R., and U54 DK107977 to B.R.). J.W. is the Virginia Murchison Linthicum Scholar in Medical Research. L.Y. was supported by a Hamon Center for Regenerative Science and Medicine fellowship from UT Southwestern Medical Center. S.P. was supported by a postdoctoral fellowship from the Deutsche Forschungsgemeinschaft (PR 1668/1-1). E.N.F. was supported by an NIH pre-doctoral training grant (5T32HL007444-35). M.L.A. was supported by an NIH training grant (T32GM008806-18). J.C.I.B. was supported by the Moxie Foundation.
Author information
Authors and Affiliations
Contributions
N.C.C. and B.R. designed and supervised the experiments, analysis and data interpretation. Y.Z. implemented the analysis pipeline, analyzed all of the sequencing datasets, interpreted the results and designed the experiments for the HERV-H functional studies. T.L. generated the CRISPR–Cas9-edited cell lines for the HERV-H functional studies, and performed differentiation and quantitative PCR of the corresponding cell lines. S.P. performed the Hi-C experiments for all stages of the cardiomyocyte differentiation, and helped with interpretation of the results. M.L.A. analyzed the HERV-H knock-in data with help from Y.Q. regarding allelic analysis. J.D.G. and E.N.F. performed the cell culture and differentiation, and collected the cells for the Hi-C, ChIP-Seq and RNA-Seq assays. E.D. contributed to analysis and interpretation of the ChIP-Seq data. R.H. performed the Hi-C experiments for the HERV-H knockout, CRISPRi, HERV-H knock-in and primate iPSC cell lines. The ChIP-Seq experiments were performed by A.Y.L. (H3K27ac), S.C. (CTCF), and Q.Z. and H.H. (SMC3). Y.Q. and R.F. helped with the analysis of the Hi-C datasets. K.M. helped with the genome editing experiments. L.Y., J.C.I.B. and J.W. cultured and prepared the non-human primate iPSCs for sequencing and interpreted the data. Z.Y. performed the RNA-Seq experiments. S.M.E. helped with interpretation of the results. Y.Z., T.L., S.P., N.C.C. and B.R. wrote the manuscript with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
B.R. is a co-founder of Arima Genomics.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Imaging and Flow cytometry data for cardiomyocyte differentiation.
(a) Immunostaining for MYL2 (white) shows exclusive expression in MYL2-H2B-GFP + hESC-derived cardiomyocytes after 80 days of cardiac differentiation. MYL2-H2B-GFP + cells were positive for cardiac troponin T (cTnT) (red), but not all cTnT+ cells were MYL2+ cardiomyocytes. DNA was stained with DAPI (blue). Images are representative of a minimum of three independent experiments. (b) Flow cytometric quantification of distinct markers for D2-D15 time points: T (D2), KDR/PDGFRα (D5), cTnT (D15). H2B-GFP + ventricular cardiomyocytes were sorted at day 80. Numbers represent the respective percentage of cells. The experiments were routinely carried out in Chi lab with similar results and had been repeated two times in this study.
Supplementary Figure 2 Quality metrics and reproducibility of sequencing datasets.
(a) Smoothed scatterplot of the Hi-C contacts between biological replicates for each stage. bin size = 100 kb. Spearman correlation coefficients are indicated on top of each sample pair. (b) Hierarchical clustering of Hi-C contact matrices based on 1-PCC (Spearman correlation coefficients) of the contacts between all samples, bin size = 100 kb. (c) Hierarchical clustering of the compartment A/B scores (PC1 values) between samples. (d) Hierarchical clustering of the insulation scores between samples. (e) Boxplot showing the number of reads for each sample. (f) Boxplot showing the percentage of alignments to human genome for each sample. (g) Boxplot showing the percentage of potential duplicated reads for each sample. (h) Boxplot showing the number of peaks called for each ChIP-seq sample. (e-h) For each boxplot the sample size N=12, the elements of the boxplot are: center line, median; box limit, upper and lower quartiles; whiskers, 1.5x interquartile range. (i) Barplots showing the relative expression levels (by RNA-seq) of representative genes during cardiomyocyte differentiation.
Supplementary Figure 3 Global changes in chromatin organization during cardiomyocyte differentiation.
(a) Frequency distribution of Hi-C contacts over genomic distances (log2) at different stages of cardiomyocyte differentiation (colored lines). (b) Barplot showing the percentages of compartment switches for each stage transition. (c) A histogram showing the distribution of the Pearson correlation coefficients (PCC) between gene expression levels and PC1 (indicating compartment A or B) value derived from the Hi-C contact matrix. (d) Snapshots of genome browser view of SOX2, HAND2 and RYR2 loci, showing PC1 values (Blue/red), RNA-seq (Black) and H3K27ac (Blue) signals.
Supplementary Figure 4 Dynamics of TADs during cardiomyocyte differentiation.
(a) Number of TADs in each stage of differentiation as defined by a domain caller algorithm first reported by Dixon et al. 20125. (b) Number of TADs (or loop domains) found by Arrowhead algorithm10. (c) Number of TADs found by Insulation score29. (d) Fraction of TAD boundaries that contain CTCF ChIP-seq peaks at each stage of cardiomyocyte differentiation. ‘stable’ group stands for TADs that are present at all stages. (e) Boxplot showing the dynamics of H3K27ac signal at ESC(+) TADs (N=198). (f) Boxplot showing the dynamics of RNA-seq signal at ESC(+) TADs (N=198). The elements of the boxplot are: center line, median; box limit, upper and lower quartiles; whiskers, 1.5x interquartile range.
Supplementary Figure 5 HERV-H elements are enriched at ESC(+) TAD boundaries.
(a) Aggregated RNA-seq expression profile (RPKM normalized) at ESC(+) TAD boundaries that overlap each of the eight repeat elements (HERVH-int and LTR7 are combined as they both belong to HERV-H). (b) Heatmap of DI scores within 70 kb of the top 50 most highly-expressed HERV-Hs. (c,d,e) heatmaps displaying Hi-C contacts for three ESC(+) TAD boundaries harboring HERV-H/LTR7 sequences are shown together with the DI, CTCF ChIP-seq, histone modification H3K27ac and RNA-seq profiles at the corresponding regions.
Supplementary Figure 6 Further characterization of the relationship between transcription levels at HERV-H elements and regular genes and formation of TAD boundaries.
(a) Aggregated DI score profiles of the TAD-boundary associated with HERV-Hs in multiple H1 ESC derived lineages and iPSCs. ESC: embryonic stem cell; MES: mesendoderm. MSC: mesenchymal stem cell; NPC: neural progenitor cell; TRO: trophoblast-like cell; iPSC: induced pluripotent stem cell. Interestingly, in human mesendoderm cells (an early human embryonic state that gives rise to mesoderm and endoderm cells), both the expression of these HERV-Hs and their corresponding TAD boundary strength were approximately half the levels compared to those observed in hESCs. There might be some un-differentiated cells in this population. (b) Heatmaps showing the aggregated DI score profile centered on the TSSs of genes (ranked by expression levels from high to low; every 1000 genes were segregated into bins, and separated by whether or not having a CTCF peak within 20kb of TSS). The DI score profile of top 50 HERV-H is shown at the bottom and the distribution of the HERV-H’s rank is shown on the right.
Supplementary Figure 7 Transcription of a solo LTR7 is correlated with appearance of TAD boundary in primate PSCs but not in mouse ESC.
(a) Hi-C interaction matrices of a solo LTR7 loci located at ESC(+) TAD boundaries at D0, D2 and D5 (top) are shown as heatmaps along with genome browser tracks of DI score, POLR2A, SMC3, CTCF, H3K27ac ChIP-seq and RNA-seq data of the expanded genomic region containing the TAD boundary (arrow). (b) Hi-C interaction matrices of the syntenic regions in bonobo iPSC, chimp iPSC and mouse ESC. (marmoset data not shown because the syntenic region is in an unassembled contig).
Supplementary Figure 8 Enrichment of TF or histone ChIP-seq signals at TAD associated HERV-H loci.
Barplot shows the ChIP-seq signal fold enrichment of the top 50 HERV-Hs comparing to HERV-Hs ranking 51-300. The red dashed line (value = 1) shows no fold enrichment.
Supplementary Figure 9 HERV-H knockout leads to alterations of gene expression programs in hESCs.
(a) Boxplots showing expression levels (RPKMs) of genes whose TSSs are located within TADs immediately 5’ (N=43) or 3’ (N=28) to boundary-associated HERV-Hs. P-values are from two-sided paired t-test on the log-transformed expression levels. The elements of the boxplot are: center line, median; box limit, upper and lower quartiles; whiskers, 1.5x interquartile range. (b) MA-plot (log ratio vs mean) showing average gene expression levels and fold changes of each gene in HERV-H1-KO and wild-type (WT). (c) Same as (b) but for HERV-H2-KO. (d) Scatterplot shows the changes in gene expression in HERV-H1-KO and HERV-H2-KO cells over WT cells. The red dots mark genes that with significantly changed gene expression in both mutant cell lines. The numbers of significantly changed genes in each Quadrant are indicated at the corner of each quadrant. Pearson correlation coefficient (PCC) and p-value are indicated (total number of genes N= 15623). (e) Barplot showing the number of significantly changed genes located within 20 kb of the HERV-H sequences. Genes down-regulated in both HERV-H knockouts were more likely to be within 20 kb of HERV-H sequences. P-value is from two-sided Fisher’s exact test (N=76). (f,g) RNA-seq profile of wild-type (WT) and HERV-H1-KO and HERV-H2-KO lines at the SCGB3A2 and LINC00458/HBL1 gene loci. The experiments were repeated twice independently with similar results.
Supplementary Figure 10 Analysis of chromatin contacts between actively transcribed HERV-Hs.
(a) Heatmap showing the contact matrix between the HERV-H1 locus and LINC00458/HBL1 locus. There is no visible chromatin contact between the two loci. (b) Heatmap shows the averaged and normalized (scaled and centered to zero) contact frequencies among each pair of HERV-H loci with RPKM greater than 1 (N=122). X-axis and y-axis show the genomic regions ±40kb surrounding HERV-H.
Supplementary Figure 11 Characterization of de novo HERV-H insertions in two engineered human ESC clones.
(a) Chromosomal view of de novo HERV-H insertions in the HERV-H-ins.clone1 transgenic line. The y-axis shows the counts of Hi-C read pairs with one end mapped to the HERV-H2 sequence. Based on proximity ligation principle, loci with high pileup should harbor HERV-H2 insertion. (b) Same as (a) but for the HERV-H-ins.clone2 transgenic line. (c) Genome browser view showing the Directionality Index (DI) score and transcription levels (RPKM) of the parental and HERV-H inserted cell lines. Note that in HERV-H-ins.clone1, the predicted HERV-H insertion creates a chimeric transcript with PIWIL1, which is not expressed in the other two cell lines. The experiments were repeated twice independently with similar results.
Supplementary Figure 12 Analysis of HERV-H related chromatin architecture and LTR7s in different primate PSC lines and in the mouse ESC.
(a) Heatmaps of DI scores within 70 kb of the syntenic regions of top 50 most highly-expressed human HERV-Hs in other indicated species. (b) Bar graph shows the percentage of HERV-Hs flanked by various types of LTRs. HERV-Hs are ranked by their expression levels in the hESCs, and grouped by bins of 50. (c) Violin plot shows the length of the flanking LTRs. HERV-Hs ranked and binned (N=50) as described in (b). The violin box displays kernel density of the LTR length distribution. (d) Boxplot shows the sequence divergence of the 5′ LTR and 3′ LTR for each bin of HERV-Hs. HERV-Hs ranked and binned (N=50) same as (b). The elements of the boxplot are: center line, median; box limit, upper and lower quartiles; whiskers, 1.5x interquartile range.
Supplementary Figure 13 A working model for HERV-H mediated TAD boundary formation.
When HERV-H is highly transcribed, the RNA polymerase complex moves directionally from the 5′LTR towards 3′LTR. Accumulation of the RNA polymerase complex presents a significant physical barrier to the movement of the cohesin complex (shown as a ring), causing it to accumulate at the 3′ end of HERV-H sequences, leading to creation of the TAD boundary. A previously characterized enhancer42 located at the 5′ LTR may regulate genes on its 5′ TAD, while the enhancer function is blocked by the TAD boundary formed downstream of the 3′ LTR.
Supplementary information
Supplementary Information
Supplementary Figs. 1–13
Supplementary Table 1
Summary statistics for the Hi-C data.
Supplementary Table 2
List of stage-specific TAD boundaries.
Supplementary Table 3
List of differentially expressed genes in HERV-H1-KO and HERV-H2-KO.
Supplementary Table 4
List of HERV-H insertions.
Supplementary Table 5
List of the primers and cell lines used in this study.
Rights and permissions
About this article
Cite this article
Zhang, Y., Li, T., Preissl, S. et al. Transcriptionally active HERV-H retrotransposons demarcate topologically associating domains in human pluripotent stem cells. Nat Genet 51, 1380–1388 (2019). https://doi.org/10.1038/s41588-019-0479-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-019-0479-7
This article is cited by
-
CRISPR-TE: a web-based tool to generate single guide RNAs targeting transposable elements
Mobile DNA (2024)
-
Ribosomal profiling of human endogenous retroviruses in healthy tissues
BMC Genomics (2024)
-
Cell-type differential targeting of SETDB1 prevents aberrant CTCF binding, chromatin looping, and cis-regulatory interactions
Nature Communications (2024)
-
Implications of the three-dimensional chromatin organization for genome evolution in a fungal plant pathogen
Nature Communications (2024)
-
Towards targeting transposable elements for cancer therapy
Nature Reviews Cancer (2024)