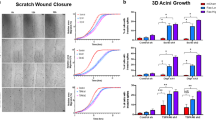
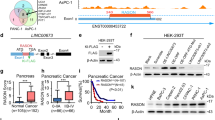
Abstract
Pancreatic ductal adenocarcinoma is an aggressive cancer with limited treatment options1. Approximately 10% of cases exhibit familial predisposition, but causative genes are not known in most families2. We perform whole-genome sequence analysis in a family with multiple cases of pancreatic ductal adenocarcinoma and identify a germline truncating mutation in the member of the RAS oncogene family-like 3 (RABL3) gene. Heterozygous rabl3 mutant zebrafish show increased susceptibility to cancer formation. Transcriptomic and mass spectrometry approaches implicate RABL3 in RAS pathway regulation and identify an interaction with RAP1GDS1 (SmgGDS), a chaperone regulating prenylation of RAS GTPases3. Indeed, the truncated mutant RABL3 protein accelerates KRAS prenylation and requires RAS proteins to promote cell proliferation. Finally, evidence in patient cohorts with developmental disorders implicates germline RABL3 mutations in RASopathy syndromes. Our studies identify RABL3 mutations as a target for genetic testing in cancer families and uncover a mechanism for dysregulated RAS activity in development and cancer.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Howlader, N. et al. SEER cancer statistics review, 1975-2011. National Cancer Institute http://seer.cancer.gov/csr/1975_2011/ (2014).
Rustgi, A. K. Familial pancreatic cancer: genetic advances. Genes Dev. 28, 1–7 (2014).
Williams, C. L. A new signaling paradigm to control the prenylation and trafficking of small GTPases. Cell Cycle 12, 2933–2934 (2013).
Chopra, S. S. et al. Inherited CHST11/MIR3922 deletion is associated with a novel recessive syndrome presenting with skeletal malformation and malignant lymphoproliferative disease. Mol. Genet. Genom. Med. 3, 413–423 (2015).
Berghmans, S. et al. tp53 mutant zebrafish develop malignant peripheral nerve sheath tumors. Proc. Natl Acad. Sci. USA 102, 407–412 (2005).
Spitsbergen, J. M. et al. Neoplasia in zebrafish (Danio rerio) treated with 7,12-dimethylbenz[a]anthracene by two exposure routes at different developmental stages. Toxicol. Pathol. 28, 705–715 (2000).
Li, Q. et al. Evaluation of the novel gene Rabl3 in the regulation of proliferation and motility in human cancer cells. Oncol. Rep. 24, 433–440 (2010).
Sowa, M. E., Bennett, E. J., Gygi, S. P. & Harper, J. W. Defining the human deubiquitinating enzyme interaction landscape. Cell 138, 389–403 (2009).
Berg, T. J. et al. Splice variants of SmgGDS control small GTPase prenylation and membrane localization. J. Biol. Chem. 285, 35255–35266 (2010).
Schuld, N. J. et al. SmgGDS-558 regulates the cell cycle in pancreatic, non-small cell lung, and breast cancers. Cell Cycle 13, 941–952 (2014).
Ntantie, E. et al. An adenosine-mediated signaling pathway suppresses prenylation of the GTPase Rap1B and promotes cell scattering. Sci. Signal. 6, ra39 (2013).
Berndt, N., Hamilton, A. D. & Sebti, S. M. Targeting protein prenylation for cancer therapy. Nat. Rev. Cancer 11, 775–791 (2011).
Jones, S. et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 321, 1801–1806 (2008).
Waddell, N. et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 518, 495–501 (2015).
Drosten, M. et al. Genetic analysis of Ras signalling pathways in cell proliferation, migration and survival. EMBO J. 29, 1091–1104 (2010).
Rauen, K. A. The RASopathies. Annu. Rev. Genom. Hum. Genet. 14, 355–369 (2013).
Wang, W. et al. Mice lacking Nf1 in osteochondroprogenitor cells display skeletal dysplasia similar to patients with neurofibromatosis type I. Hum. Mol. Genet. 20, 3910–3924 (2011).
Hernandez-Porras, I. et al. K-RasV14I recapitulates Noonan syndrome in mice. Proc. Natl Acad. Sci. USA 111, 16395–16400 (2014).
Roberts, N. J. et al. Whole genome sequencing defines the genetic heterogeneity of familial pancreatic cancer. Cancer Discov. 6, 166–175 (2016).
Wheeler, D. B., Zoncu, R., Root, D. E., Sabatini, D. M. & Sawyers, C. L. Identification of an oncogenic RAB. Protein Sci. 350, 211–217 (2015).
Jindal, G. A. et al. In vivo severity ranking of Ras pathway mutations associated with developmental disorders. Proc. Natl Acad. Sci. USA 114, 510–515 (2017).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
DePristo, M. A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498 (2011).
Cox, A. G. et al. Yap reprograms glutamine metabolism to increase nucleotide biosynthesis and enable liver growth. Nat. Cell Biol. 18, 886–896 (2016).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Behrends, C., Sowa, M. E., Gygi, S. P. & Harper, J. W. Network organization of the human autophagy system. Nature 466, 68–76 (2010).
Huttlin, E. L. et al. The BioPlex network: a systematic exploration of the human interactome. Cell 162, 425–440 (2015).
Rauniyar, N. Parallel reaction monitoring: a targeted experiment performed using high resolution and high mass accuracy mass spectrometry. Int. J. Mol. Sci. 16, 28566–28581 (2015).
MacLean, B. et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968 (2010).
Krieger, E. et al. Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: four approaches that performed well in CASP8. Proteins 77, 114–122 (2009).
Xu, D. & Zhang, Y. Ab initio protein structure assembly using continuous structure fragments and optimized knowledge-based force field. Proteins 80, 1715–1735 (2012).
Pierce, B. G., Hourai, Y. & Weng, Z. Accelerating protein docking in ZDOCK using an advanced 3D convolution library. PLoS ONE 6, e24657 (2011).
Ouyang, H. et al. Response of immortalized murine cementoblasts/periodontal ligament cells to parathyroid hormone and parathyroid hormone-related protein in vitro. Arch. Oral. Biol. 45, 293–303 (2000).
Acknowledgements
This work was supported by the NIH grant nos. K08 DK105326 (to S.N.); R01 DK090311, R01 DK095721 and R24 OD017870 (to W.G.); R01 GM095567, R01 CA157490, R01 CA188048, P01 CA117969 and R35 CA232124 (to A.C.K.); R01 CA188871 (to C.W.) and R01 GM040602 (to C.A.F.); as well as grants from the National Pancreas Foundation (to S.N.), the Harvard Digestive Diseases Center (grant no. P30 DK034854 to S.N. and W.G.), the Ken and Louise Goldberg Award (to S.N.), an ACS Research Scholar Grant (RSG-13-298-01-TBG to A.C.K.), the Lustgarten Foundation and SU2C (to A.C.K) and the Anna Fuller Fund and the Claudia Adams Barr Program for Innovative Cancer Research (to W.G.). S.N. is a recipient of the Burroughs Wellcome Fund Career Award for Medical Scientists. W.G. is a Pew Scholar in the Biomedical Sciences.
Author information
Authors and Affiliations
Contributions
S.N. and W.G. conceived and designed the overall project. S.S. and C.I.U. assisted with selecting the family, gathering the clinical histories and collecting DNA samples under human subject IRB-approved protocols. S.N., W.G. and I.L. designed the WGS analysis. I.L. performed the WGS analysis and candidate variant filtering. S.N., J.W., A.J.K., J.E.H., A.G.C. and J.H. designed and generated the zebrafish rabl3 mutant lines and performed the cancer studies. J.R.H. and S.N. performed zebrafish histology preparation and analysis. J.D.M. performed and analyzed the AP–MS experiments and CompPASS suite protein interactomics. S.N., W.G. and C.W. conceived and designed the in vitro immunoprecipitation, prenylation assays and HEK293T cell proliferation assays, and P.G., A.B., E.L. and B.U. performed these experiments. S.N. and O.M. designed and performed RASless MEF experiments. J.W.P. performed protein structural modeling. B.C.J. and C.A.F. designed and performed purification of recombinant protein. J.A.P., S.G. and J.D.M. assisted with mass spectrometry analysis. Y.H. assisted with RNA-seq data analysis. M.B.G. performed the zebrafish µCT and bone histomorphometric analysis. O.M., X.W. and J.D.M. provided assistance with tissue culture experiments. C.A.C. and J.A.R. provided analysis of clinical exome sequencing data. C.A.C. and I.L. provided analysis of variants in the Exome Aggregation Consortium. J.W.H., G.G., S.R.S., K.C. and A.C.K. provided overall input. S.N. and W.G. wrote the manuscript. All authors reviewed and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
A.C.K. has financial interests in Vescor Therapeutics, LLC. A.C.K. is an inventor on patents pertaining to Kras-regulated metabolic pathways, redox control pathways in pancreatic cancer, targeting GOT1 as a therapeutic approach and the autophagic control of iron metabolism. A.C.K. is on the SAB of Cornerstone/Rafael Pharmaceuticals. G.G. receives research funds from IBM and Pharmacyclics. W.G. receives patent royalties from FATE Therapeutics and is on the SAB of Camp4 Therapeutics.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–19 and Supplementary Tables 3 and 5
Supplementary Table 1
Supplementary Table 1
Supplementary Table 2
Supplementary Table 2
Supplementary Table 4
Supplementary Table 4
Supplementary Video 1
Supplementary Video 1
Supplementary Video 2
Supplementary Video 2
Supplementary Video 3
Supplementary Video 3
Rights and permissions
About this article
Cite this article
Nissim, S., Leshchiner, I., Mancias, J.D. et al. Mutations in RABL3 alter KRAS prenylation and are associated with hereditary pancreatic cancer. Nat Genet 51, 1308–1314 (2019). https://doi.org/10.1038/s41588-019-0475-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-019-0475-y
This article is cited by
-
Computational analysis of cancer genome sequencing data
Nature Reviews Genetics (2022)
-
Unique roles of rare variants in the genetics of complex diseases in humans
Journal of Human Genetics (2021)
-
Zebrafish disease models in drug discovery: from preclinical modelling to clinical trials
Nature Reviews Drug Discovery (2021)
-
Knockdown of RABL3 suppresses the proliferation and invasion of oral squamous cell carcinoma through inactivating the FAK/AKT pathway
Journal of Bioenergetics and Biomembranes (2021)
-
The genetics of ductal adenocarcinoma of the pancreas in the year 2020: dramatic progress, but far to go
Modern Pathology (2020)