Abstract
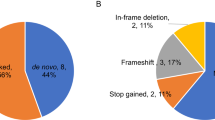
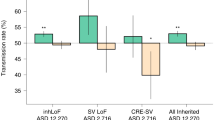
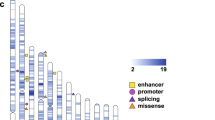
Autism spectrum disorder (ASD) affects up to 1 in 59 individuals1. Genome-wide association and large-scale sequencing studies strongly implicate both common variants2,3,4 and rare de novo variants5,6,7,8,9,10 in ASD. Recessive mutations have also been implicated11,12,13,14 but their contribution remains less well defined. Here we demonstrate an excess of biallelic loss-of-function and damaging missense mutations in a large ASD cohort, corresponding to approximately 5% of total cases, including 10% of females, consistent with a female protective effect. We document biallelic disruption of known or emerging recessive neurodevelopmental genes (CA2, DDHD1, NSUN2, PAH, RARB, ROGDI, SLC1A1, USH2A) as well as other genes not previously implicated in ASD including FEV (FEV transcription factor, ETS family member), which encodes a key regulator of the serotonergic circuitry. Our data refine estimates of the contribution of recessive mutation to ASD and suggest new paths for illuminating previously unknown biological pathways responsible for this condition.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
Data included in this manuscript have been deposited at the database of Genotypes and Phenotypes, merged with published data under accession number phs000298.v4.p3. Correspondence and requests for materials should be addressed to Timothy.yu@childrens.harvard.edu.
References
Baio, J. et al. Prevalence of autism spectrum disorder among children aged 8 years. Autism and developmental disabilities monitoring network, 11 sites, United States, 2014. MMWR Surveill. Summ. 63, 1–22 (2014).
Robinson, E. B. et al. Genetic risk for autism spectrum disorders and neuropsychiatric variation in the general population. Nat. Genet. 48, 552–555 (2016).
Gaugler, T. et al. Most genetic risk for autism resides with common variation. Nat. Genet. 46, 881–885 (2014).
Weiner, D. J. et al. Polygenic transmission disequilibrium confirms that common and rare variation act additively to create risk for autism spectrum disorders. Nat. Genet. 49, 978–985 (2017).
Krumm, N. et al. Excess of rare, inherited truncating mutations in autism. Nat. Genet. 47, 582–588 (2015).
De Rubeis, S. et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 515, 209–215 (2014).
Levy, D. et al. Rare de novo and transmitted copy-number variation in autistic spectrum disorders. Neuron 70, 886–897 (2011).
Sanders, S. J. et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature 485, 237–241 (2012).
Iossifov, I. et al. De novo gene disruptions in children on the autistic spectrum. Neuron 74, 285–299 (2012).
Ronemus, M., Iossifov, I., Levy, D. & Wigler, M. The role of de novo mutations in the genetics of autism spectrum disorders. Nat. Rev. Genet. 15, 133–141 (2014).
Morrow, E. M. et al. Identifying autism loci and genes by tracing recent shared ancestry. Science 321, 218–223 (2008).
Chahrour, M. H. et al. Whole-exome sequencing and homozygosity analysis implicate depolarization-regulated neuronal genes in autism. PLoS Genet. 8, e1002635 (2012).
Yu, T. W. et al. Using whole-exome sequencing to identify inherited causes of autism. Neuron 77, 259–273 (2013).
Lim, E. T. et al. Rare complete knockouts in humans: population distribution and significant role in autism spectrum disorders. Neuron 77, 235–242 (2013).
Buxbaum, J. D. et al. The autism sequencing consortium: large-scale, high-throughput sequencing in autism spectrum disorders. Neuron 76, 1052–1056 (2012).
Huang, N., Lee, I., Marcotte, E. M. & Hurles, M. E. Characterising and predicting haploinsufficiency in the human genome. PLoS Genet. 6, e1001154 (2010).
Lek, M. et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 (2016).
Betancur, C. Etiological heterogeneity in autism spectrum disorders: more than 100 genetic and genomic disorders and still counting. Brain Res. 1380, 42–77 (2011).
Jacquemont, S. et al. A higher mutational burden in females supports a “female protective model” in neurodevelopmental disorders. Am. J. Hum. Genet. 94, 415–425 (2014).
Robinson, E. B., Lichtenstein, P., Anckarsäter, H., Happé, F. & Ronald, A. Examining and interpreting the female protective effect against autistic behavior. Proc. Natl Acad. Sci. USA 110, 5258–5262 (2013).
Weiss, L. A. et al. Association between microdeletion and microduplication at 16p11.2 and autism. N. Engl. J. Med. 358, 667–675 (2008).
Blaker-Lee, A., Gupta, S., McCammon, J. M., De Rienzo, G. & Sive, H. Zebrafish homologs of genes within 16p11.2, a genomic region associated with brain disorders, are active during brain development, and include two deletion dosage sensor genes. Dis. Model. Mech. 5, 834–851 (2012).
Park, S. M., Littleton, J. T., Park, H. R. & Lee, J. H. Drosophila homolog of human KIF22 at the autism-linked 16p11.2 loci influences synaptic connectivity at larval neuromuscular junctions. Exp. Neurobiol. 25, 33–39 (2016).
Bailey, C. G. et al. Loss-of-function mutations in the glutamate transporter SLC1A1 cause human dicarboxylic aminoaciduria. J. Clin. Invest. 121, 446–453 (2011).
Teijema, H. L., van Gelderen, H. H., Giesberts, M. A. & Laurent de Angulo, M. S. Dicarboxylic aminoaciduria: an inborn error of glutamate and aspartate transport with metabolic implications, in combination with a hyperprolinemia. Metab. Clin. Exp. 23, 115–123 (1974).
Swarna, M., Rao, D. N. & Reddy, P. P. Dicarboxylic aminoaciduria associated with mental retardation. Hum. Genet. 82, 299–300 (1989).
Rothstein, J. D. et al. Localization of neuronal and glial glutamate transporters. Neuron 13, 713–725 (1994).
Ross, J. R., Porter, B. E., Buckley, P. T., Eberwine, J. H. & Robinson, M. B. mRNA for the EAAC1 subtype of glutamate transporter is present in neuronal dendrites in vitro and dramatically increases in vivo after a seizure. Neurochem. Int. 58, 366–375 (2011).
Nieoullon, A. et al. The neuronal excitatory amino acid transporter EAAC1/EAAT3: does it represent a major actor at the brain excitatory synapse? J. Neurochem. 98, 1007–1018 (2006).
Diamond, J. S. Neuronal glutamate transporters limit activation of NMDA receptors by neurotransmitter spillover on CA1 pyramidal cells. J. Neurosci. 21, 8328–8338 (2001).
Bianchi, M. G., Bardelli, D., Chiu, M. & Bussolati, O. Changes in the expression of the glutamate transporter EAAT3/EAAC1 in health and disease. Cell. Mol. Life Sci. 71, 2001–2015 (2014).
Stafford, M. M., Brown, M. N., Mishra, P., Stanwood, G. D. & Mathews, G. C. Glutamate spillover augments GABA synthesis and release from axodendritic synapses in rat hippocampus. Hippocampus 20, 134–144 (2010).
Mathews, G. C. & Diamond, J. S. Neuronal glutamate uptake contributes to GABA synthesis and inhibitory synaptic strength. J. Neurosci. 23, 2040–2048 (2003).
Scimemi, A., Tian, H. & Diamond, J. S. Neuronal transporters regulate glutamate clearance, NMDA receptor activation, and synaptic plasticity in the hippocampus. J. Neurosci. 29, 14581–14595 (2009).
Peghini, P., Janzen, J. & Stoffel, W. Glutamate transporter EAAC-1-deficient mice develop dicarboxylic aminoaciduria and behavioral abnormalities but no neurodegeneration. EMBO J. 16, 3822–3832 (1997).
Lee, S., Park, S. H. & Zuo, Z. Effects of isoflurane on learning and memory functions of wild-type and glutamate transporter type 3 knockout mice. J. Pharm. Pharmacol. 64, 302–307 (2012).
Lucki, I. The spectrum of behaviors influenced by serotonin. Biol. Psychiatry 44, 151–162 (1998).
Muller, C. L., Anacker, A. M. J. & Veenstra-VanderWeele, J. The serotonin system in autism spectrum disorder: from biomarker to animal models. Neuroscience 321, 24–41 (2016).
Hendricks, T., Francis, N., Fyodorov, D. & Deneris, E. S. The ETS domain factor Pet-1 is an early and precise marker of central serotonin neurons and interacts with a conserved element in serotonergic genes. J. Neurosci. 19, 10348–10356 (1999).
Hendricks, T. J. et al. Pet-1 ETS gene plays a critical role in 5-HT neuron development and is required for normal anxiety-like and aggressive behavior. Neuron 37, 233–247 (2003).
Liu, C. et al. Pet-1 is required across different stages of life to regulate serotonergic function. Nat. Neurosci. 13, 1190–1198 (2010).
Iyo, A. H., Porter, B., Deneris, E. S. & Austin, M. C. Regional distribution and cellular localization of the ETS-domain transcription factor, FEV, mRNA in the human postmortem brain. Synapse 57, 223–228 (2005).
Maurer, P. et al. The Ets transcription factor Fev is specifically expressed in the human central serotonergic neurons. Neurosci. Lett. 357, 215–218 (2004).
Cao, H. et al. FCHSD1 and FCHSD2 are expressed in hair cell stereocilia and cuticular plate and regulate actin polymerization in vitro. PLoS ONE 8, e56516 (2013).
Shao, W., Halachmi, S. & Brown, M. ERAP140, a conserved tissue-specific nuclear receptor coactivator. Mol. Cell. Biol. 22, 3358–3372 (2002).
Poon, C. L. C., Mitchell, K. A., Kondo, S., Cheng, L. Y. & Harvey, K. F. The Hippo pathway regulates neuroblasts and brain size in Drosophila melanogaster. Curr. Biol. 26, 1034–1042 (2016).
Cheng, Y. Z. et al. Investigating embryonic expression patterns and evolution of AHI1 and CEP290 genes, implicated in Joubert syndrome. PLoS ONE 7, e44975 (2012).
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010).
Stenson, P. D. et al. The Human Gene Mutation Database: towards a comprehensive repository of inherited mutation data for medical research, genetic diagnosis and next-generation sequencing studies. Hum. Genet. 136, 665–677 (2017).
Ng, P. C. & Henikoff, S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 31, 3812–3814 (2003).
Adzhubei, I. A. et al. A method and server for predicting damaging missense mutations. Nat. Methods 7, 248–249 (2010).
Schwarz, J. M., Rödelsperger, C., Schuelke, M. & Seelow, D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat. Methods 7, 575–576 (2010).
Reva, B., Antipin, Y. & Sander, C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 39, e118 (2011).
Chun, S. & Fay, J. C. Identification of deleterious mutations within three human genomes. Genome Res. 19, 1553–1561 (2009).
Browning, S. R. & Browning, B. L. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am. J. Hum. Genet. 81, 1084–1097 (2007).
Gerrelli, D., Lisgo, S., Copp, A. J. & Lindsay, S. Enabling research with human embryonic and fetal tissue resources. Development 142, 3073–3076 (2015).
Kerwin, J. et al. The HUDSEN Atlas: a three-dimensional (3D) spatial framework for studying gene expression in the developing human brain. J. Anat. 217, 289–299 (2010).
Acknowledgements
We thank A. Hossain and N. Hatem for their help with sample preparation, and J. Kerwin for her help with analysis of in situ hybridization results. R.N.D. was supported by an NIH T32 fellowship from the Fundamental Neurobiology Training Grant (no. 5 T32 NS007484-14) and the Nancy Lurie Marks Clinical and Research Fellowship Program in Autism. The ASC is supported by the National Institute of Mental Health (NIMH; grant nos. MH100233, MH100229, MH100209, MH100239, MH111661, MH111660, MH111662 and MH111658). Collection of the PAGES cohort is supported by the NIMH (grant no. MH097849). This work was supported in part through the computational resources provided by the Department of Scientific Computing at the Icahn School of Medicine at Mount Sinai, and the Research Information Technology Group at Harvard Medical School, which is partially supported by National Institutes of Health grant no. NCRR 1S10RR028832-01. Human embryonic and fetal material was provided jointly by the MRC/Wellcome Trust (grant no. MR/R006237/1) Human Developmental Biology Resource (www.hdbr.org). C.A.W. is an Investigator of the Howard Hughes Medical Institute. C.A.W. and T.W.Y. were supported by NIMH grant nos. RC2MH089952 and RO1MH083565. T.W.Y. was supported by grant nos. NIH/NIMH R01MH113761, NICHD/NHGRI/NIH U19HD077671 and NIH/NICHD U24HD0938487, and by a SFARI Pilot Research Award. S.D.R. and J.D.B. are supported by the Beatrice and Samuel A. Seaver Foundation.
Author information
Authors and Affiliations
Consortia
Contributions
T.W.Y., R.N.D., E.T.L. and M.J.D. designed the study, with important additional contributions from C.B., D.J.C, C.A.W. and J.D.B. R.N.D., E.T.L. and A.S. performed the data analyses. S.D.R. and S.G. performed the Sanger validation. A.G.C. and C.M.F. characterized the FEV family. L.O., S.G. and T.W.Y. designed and performed the in situ expression analyses. R.N.D. and T.W.Y. wrote the manuscript. All authors reviewed and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–11 and Supplementary Note
Supplementary Tables
Supplementary Tables 1–13
Rights and permissions
About this article
Cite this article
Doan, R.N., Lim, E.T., De Rubeis, S. et al. Recessive gene disruptions in autism spectrum disorder. Nat Genet 51, 1092–1098 (2019). https://doi.org/10.1038/s41588-019-0433-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-019-0433-8
This article is cited by
-
Pathogenic/likely pathogenic mutations identified in Vietnamese children diagnosed with autism spectrum disorder using high-resolution SNP genotyping platform
Scientific Reports (2024)
-
Comparative yield of molecular diagnostic algorithms for autism spectrum disorder diagnosis in India: evidence supporting whole exome sequencing as first tier test
BMC Neurology (2023)
-
Transmembrane protein 184B (TMEM184B) promotes expression of synaptic gene networks in the mouse hippocampus
BMC Genomics (2023)
-
Sex differences in the transcriptome of extracellular vesicles secreted by fetal neural stem cells and effects of chronic alcohol exposure
Biology of Sex Differences (2023)
-
Comparison of three bioinformatics tools in the detection of ASD candidate variants from whole exome sequencing data
Scientific Reports (2023)