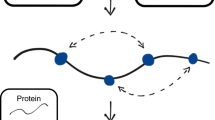
Abstract
Determining the three-dimensional structures of macromolecules is a major goal of biological research, because of the close relationship between structure and function; however, thousands of protein domains still have unknown structures. Structure determination usually relies on physical techniques including X-ray crystallography, NMR spectroscopy and cryo-electron microscopy. Here we present a method that allows the high-resolution three-dimensional backbone structure of a biological macromolecule to be determined only from measurements of the activity of mutant variants of the molecule. This genetic approach to structure determination relies on the quantification of genetic interactions (epistasis) between mutations and the discrimination of direct from indirect interactions. This provides an alternative experimental strategy for structure determination, with the potential to reveal functional and in vivo structures.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
No primary data were generated in this study. Data sources are listed in the Methods at appropriate places. Processed interaction scores for all datasets are included in Supplementary Table 1. All intermediate steps of data processing can be recapitulated with the scripts provided at https://github.com/lehner-lab/DMS2structure.
Code availability
Paired-end sequencing reads were merged with USearch v.10.0.240. Data were analyzed with custom scripts written and executed in the R programming language, v.3.4.3. Structural simulations were performed with Xplor-NIH modeling suite v.2.46. TM-Score72 (update 23 March 2016) was used to evaluate accuracy of structural models. PSIPRED v.3.3 was used to predict secondary structure elements from amino acid sequences. PyMOL v.1.8.6.073 was used to visualize protein structures. All custom scripts needed to repeat the analyses are available at https://github.com/lehner-lab/DMS2structure.
References
Ovchinnikov, S. et al. Protein structure determination using metagenome sequence data. Science 355, 294–298 (2017).
Tokuriki, N. & Tawfik, D. S. Stability effects of mutations and protein evolvability. Curr. Opin. Struct. Biol. 19, 596–604 (2009).
Lehner, B. Molecular mechanisms of epistasis within and between genes. Trends Genet. 27, 323–331 (2011).
Fowler, D. M. & Fields, S. Deep mutational scanning: a new style of protein science. Nat. Methods 11, 801–807 (2014).
Starr, T. N. & Thornton, J. W. Epistasis in protein evolution. Protein Sci. 25, 1204–1218 (2016).
Horovitz, A. & Fersht, A. R. Strategy for analysing the co-operativity of intramolecular interactions in peptides and proteins. J. Mol. Biol. 214, 613–617 (1990).
Carter, P. J., Winter, G., Wilkinson, A. J. & Fersht, A. R. The use of double mutants to detect structural changes in the active site of the tyrosyl-tRNA synthetase (Bacillus stearothermophilus). Cell 38, 835–840 (1984).
Ackermann, E. J., Ang, E. T., Kanter, J. R., Tsigelny, I. & Taylor, P. Identification of pairwise interactions in the α-neurotoxin–nicotinic acetylcholine receptor complex through double mutant cycles. J. Biol. Chem. 273, 10958–10964 (1998).
Chen, J. & Stites, W. E. Energetics of side chain packing in staphylococcal nuclease assessed by systematic double mutant cycles. Biochemistry 40, 14004–14011 (2001).
Roisman, L. C., Piehler, J., Trosset, J. Y., Scheraga, H. A. & Schreiber, G. Structure of the interferon–receptor complex determined by distance constraints from double-mutant cycles and flexible docking. Proc. Natl Acad. Sci. USA 98, 13231–13236 (2001).
Diss, G. & Lehner, B. The genetic landscape of a physical interaction. eLife 7, e32472 (2018).
Melamed, D., Young, D. L., Gamble, C. E., Miller, C. R. & Fields, S. Deep mutational scanning of an RRM domain of the Saccharomyces cerevisiae poly(A)-binding protein. RNA 19, 1537–1551 (2013).
Olson, C. A., Wu, N. C. & Sun, R. A comprehensive biophysical description of pairwise epistasis throughout an entire protein domain. Curr. Biol. 24, 2643–2651 (2014).
Sahoo, A., Khare, S., Devanarayanan, S., Jain, P. C. & Varadarajan, R. Residue proximity information and protein model discrimination using saturation-suppressor mutagenesis. eLife 4, e09532 (2015).
Li, C. & Zhang, J. Multi-environment fitness landscapes of a tRNA gene. Nat. Ecol. Evol. 2, 1025–1032 (2018).
Li, C., Qian, W., Maclean, C. J. & Zhang, J. The fitness landscape of a tRNA gene. Science 352, 837–840 (2016).
Domingo, J., Diss, G. & Lehner, B. Pairwise and higher-order genetic interactions during the evolution of a tRNA. Nature 558, 117–121 (2018).
Puchta, O. et al. Network of epistatic interactions within a yeast snoRNA. Science 352, 840–844 (2016).
Göbel, U., Sander, C., Schneider, R. & Valencia, A. Correlated mutations and residue contacts in proteins. Proteins 18, 309–317 (1994).
Altschuh, D., Lesk, A. M., Bloomer, A. C. & Klug, A. Correlation of co-ordinated amino acid substitutions with function in viruses related to tobacco mosaic virus. J. Mol. Biol. 193, 693–707 (1987).
Gloor, G. B., Martin, L. C., Wahl, L. M. & Dunn, S. D. Mutual information in protein multiple sequence alignments reveals two classes of coevolving positions. Biochemistry 44, 7156–7165 (2005).
Halabi, N., Rivoire, O., Leibler, S. & Ranganathan, R. Protein sectors: evolutionary units of three-dimensional structure. Cell 138, 774–786 (2009).
Lockless, S. W. & Ranganathan, R. Evolutionarily conserved pathways of energetic connectivity in protein families. Science 286, 295–299 (1999).
Morcos, F. et al. Direct-coupling analysis of residue coevolution captures native contacts across many protein families. Proc. Natl Acad. Sci. USA 108, E1293–E1301 (2011).
Weigt, M., White, R. A., Szurmant, H., Hoch, J. A. & Hwa, T. Identification of direct residue contacts in protein–protein interaction by message passing. Proc. Natl Acad. Sci. USA 106, 67–72 (2009).
Burger, L. & van Nimwegen, E. Disentangling direct from indirect co-evolution of residues in protein alignments. PLoS Comput. Biol. 6, e1000633 (2010).
Weinreb, C. et al. 3D RNA and functional interactions from evolutionary couplings. Cell 165, 963–975 (2016).
Tóth-Petróczy, A. et al. Structured states of disordered proteins from genomic sequences. Cell 167, 158–170 (2016).
Hopf, T. A. et al. Three-dimensional structures of membrane proteins from genomic sequencing. Cell 149, 1607–1621 (2012).
Marks, D. S. et al. Protein 3D structure computed from evolutionary sequence variation. PLoS ONE 6, e28766 (2011).
Jones, D. T., Buchan, D. W. A., Cozzetto, D. & Pontil, M. PSICOV: precise structural contact prediction using sparse inverse covariance estimation on large multiple sequence alignments. Bioinformatics 28, 184–190 (2012).
De Leonardis, E. et al. Direct-coupling analysis of nucleotide coevolution facilitates RNA secondary and tertiary structure prediction. Nucleic Acids Res. 43, 10444–10455 (2015).
Sułkowska, J. I., Morcos, F., Weigt, M., Hwa, T. & Onuchic, J. N. Genomics-aided structure prediction. Proc. Natl Acad. Sci. USA 109, 10340–10345 (2012).
Ovchinnikov, S. et al. Large-scale determination of previously unsolved protein structures using evolutionary information. eLife 4, e09248 (2015).
Ovchinnikov, S., Kamisetty, H. & Baker, D. Robust and accurate prediction of residue–residue interactions across protein interfaces using evolutionary information. eLife 3, e02030 (2014).
Matreyek, K. A. et al. Multiplex assessment of protein variant abundance by massively parallel sequencing. Nat. Genet. 50, 874–882 (2018).
Weile, J. et al. A framework for exhaustively mapping functional missense variants. Mol. Syst. Biol. 13, 957 (2017).
Rocklin, G. J. et al. Global analysis of protein folding using massively parallel design, synthesis, and testing. Science 357, 168–175 (2017).
Kim, I., Miller, C. R., Young, D. L. & Fields, S. High-throughput analysis of in vivo protein stability. Mol. Cell Proteomics 12, 3370–3378 (2013).
Marks, D. S., Hopf, T. A. & Sander, C. Protein structure prediction from sequence variation. Nat. Biotechnol. 30, 1072–1080 (2012).
Andreani, J. & Söding, J. bbcontacts: prediction of β-strand pairing from direct coupling patterns. Bioinformatics 31, 1729–1737 (2015).
Schwieters, C. D., Kuszewski, J. J., Tjandra, N. & Clore, G. M. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 160, 65–73 (2003).
Araya, C. L. et al. A fundamental protein property, thermodynamic stability, revealed solely from large-scale measurements of protein function. Proc. Natl Acad. Sci. USA 109, 16858–16863 (2012).
Liu, Y., Palmedo, P., Ye, Q., Berger, B. & Peng, J. Enhancing evolutionary couplings with deep convolutional neural networks. Cell Syst. 6, 65–74 (2018).
Schaarschmidt, J., Monastyrskyy, B., Kryshtafovych, A. & Bonvin, A. M. J. J. Assessment of contact predictions in CASP12: co-evolution and deep learning coming of age. Proteins 86, 51–66 (2018).
Fox, N. K., Brenner, S. E. & Chandonia, J. M. SCOPe: structural classification of proteins—extended, integrating SCOP and ASTRAL data and classification of new structures. Nucleic Acids Res 42, D304–D309 (2014).
Rollins, N. J. et al. Inferring protein 3D structure from deep mutation scans. Nat. Genet. https://doi.org/10.1038/s41588-019-0432-9 (2019).
Jones, D. T., Singh, T., Kosciolek, T. & Tetchner, S. MetaPSICOV: combining coevolution methods for accurate prediction of contacts and long range hydrogen bonding in proteins. Bioinformatics 31, 999–1006 (2015).
Wang, S., Sun, S., Li, Z., Zhang, R. & Xu, J. Accurate de novo prediction of protein contact map by ultra-deep learning model. PLoS Comput. Biol. 13, e1005324 (2017).
Rohl, C. A., Strauss, C. E., Misura, K. M. & Baker, D. Protein structure prediction using Rosetta. Methods Enzym. 383, 66–93 (2004).
Yang, J. et al. The I-TASSER suite: protein structure and function prediction. Nat. Methods 12, 7–8 (2015).
Poelwijk, F. J., Socolich, M. & Ranganathan, R. Learning the pattern of epistasis linking genotype and phenotype in a protein. Preprint at bioRxiv https://doi.org/10.1101/213835 (2017).
Firnberg, E. & Ostermeier, M. PFunkel: efficient, expansive, user-defined mutagenesis. PLoS ONE 7, e52031 (2012).
Wrenbeck, E. E. et al. Plasmid-based one-pot saturation mutagenesis. Nat. Methods 13, 928–930 (2016).
Starita, L. M. et al. Massively parallel functional analysis of BRCA1 RING domain variants. Genetics 200, 413–422 (2015).
Starita, L. M. et al. Activity-enhancing mutations in an E3 ubiquitin ligase identified by high-throughput mutagenesis. Proc. Natl Acad. Sci. USA 110, E1263–E1272 (2013).
Starr, T. N., Picton, L. K. & Thornton, J. W. Alternative evolutionary histories in the sequence space of an ancient protein. Nature 549, 409–413 (2017).
Fowler, D. M. et al. High-resolution mapping of protein sequence–function relationships. Nat. Methods 7, 741–746 (2010).
McLaughlin, R. N. Jr, Poelwijk, F. J., Raman, A., Gosal, W. S. & Ranganathan, R. The spatial architecture of protein function and adaptation. Nature 491, 138–142 (2012).
Bolognesi, B. et al. The mutational landscape of a prion-like domain. Preprint at bioRxiv https://doi.org/10.1101/592121 (2019).
Gallagher, T., Alexander, P., Bryan, P. & Gilliland, G. L. Two crystal structures of the B1 immunoglobulin-binding domain of streptococcal protein G and comparison with NMR. Biochemistry 33, 4721–4729 (1994).
Jones, D. T. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292, 195–202 (1999).
Rubin, A. F. et al. A statistical framework for analyzing deep mutational scanning data. Genome Biol. 18, 741 (2017).
Edgar, R. C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461 (2010).
Barlow, R. Statistics: A Guide to the Use of Statistical Methods in the Physical Sciences (Wiley, 1989).
Schäfer, J. & Strimmer, K. A shrinkage approach to large-scale covariance matrix estimation and implications for functional genomics. Stat. Appl. Genet. Mol. Biol. 4, 32 (2005).
Stein, R. R., Marks, D. S. & Sander, C. Inferring pairwise interactions from biological data using maximum-entropy probability models. PLoS Comput. Biol. 11, e1004182 (2015).
Pires, J. R. et al. Solution structures of the YAP65 WW domain and the variant L30 K in complex with the peptides GTPPPPYTVG, N-(n-octyl)-GPPPY and PLPPY and the application of peptide libraries reveal a minimal binding epitope. J. Mol. Biol. 314, 1147–1156 (2001).
Deo, R. C., Bonanno, J. B., Sonenberg, N. & Burley, S. K. Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell 98, 835–845 (1999).
Glover, J. N. & Harrison, S. C. Crystal structure of the heterodimeric bZIP transcription factor c-Fos–c-Jun bound to DNA. Nature 373, 257–261 (1995).
Seemayer, S., Gruber, M. & Söding, J. CCMpred—fast and precise prediction of protein residue–residue contacts from correlated mutations. Bioinformatics 30, 3128–3130 (2014).
Zhang, Y. & Skolnick, J. Scoring function for automated assessment of protein structure template quality. Proteins 57, 702–710 (2004).
The PyMOL Molecular Graphics System v.1.8 (Schrödinger LLC).
Acknowledgements
We thank Y. Liu and J. Peng for making their DeepContact code available and for their advice; members of the Lehner laboratory, T. Gross, G. Mönke, M. Bolognesi and C. Camilloni for discussions and feedback. This work was supported by a European Research Council (ERC) Consolidator grant (616434), the Spanish Ministry of Economy, Industry and Competitiveness (MEIC; BFU2017-89488-P), the AXA Research Fund, the Bettencourt Schueller Foundation, Agencia de Gestio d’Ajuts Universitaris i de Recerca (AGAUR, 2017 SGR 1322), the EMBL-CRG Systems Biology Program and the CERCA Program/Generalitat de Catalunya. J.M.S. was supported by an EMBO Long-Term Fellowship (ALTF 857-2016). This project has received funding from the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement 752809 (J.M.S.). We acknowledge support from the Spanish Ministry of Economy, Industry and Competitiveness (MEIC) to the EMBL partnership and the Centro de Excelencia Severo Ochoa.
Author information
Authors and Affiliations
Contributions
J.M.S. and B.L. conceptualized the study; J.M.S. developed the methods and carried out the study; J.M.S. and B.L. wrote the paper; B.L. supervised the study.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figures 1–9 and Supplementary Note
Rights and permissions
About this article
Cite this article
Schmiedel, J.M., Lehner, B. Determining protein structures using deep mutagenesis. Nat Genet 51, 1177–1186 (2019). https://doi.org/10.1038/s41588-019-0431-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-019-0431-x
This article is cited by
-
Protein destabilization underlies pathogenic missense mutations in ARID1B
Nature Structural & Molecular Biology (2024)
-
Protein design using structure-based residue preferences
Nature Communications (2024)
-
An Atlas of Variant Effects to understand the genome at nucleotide resolution
Genome Biology (2023)
-
ACIDES: on-line monitoring of forward genetic screens for protein engineering
Nature Communications (2023)
-
A computational toolbox for the assembly yield of complex and heterogeneous structures
Nature Communications (2023)