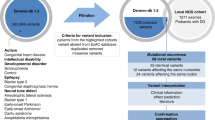
Abstract
We address the challenge of detecting the contribution of noncoding mutations to disease with a deep-learning-based framework that predicts the specific regulatory effects and the deleterious impact of genetic variants. Applying this framework to 1,790 autism spectrum disorder (ASD) simplex families reveals a role in disease for noncoding mutations—ASD probands harbor both transcriptional- and post-transcriptional-regulation-disrupting de novo mutations of significantly higher functional impact than those in unaffected siblings. Further analysis suggests involvement of noncoding mutations in synaptic transmission and neuronal development and, taken together with previous studies, reveals a convergent genetic landscape of coding and noncoding mutations in ASD. We demonstrate that sequences carrying prioritized mutations identified in probands possess allele-specific regulatory activity, and we highlight a link between noncoding mutations and heterogeneity in the IQ of ASD probands. Our predictive genomics framework illuminates the role of noncoding mutations in ASD and prioritizes mutations with high impact for further study, and is broadly applicable to complex human diseases.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
ASD WGS data can be obtained from the Simons Foundation Autism Research Initiative (SFARI). All variant predicted scores have been made available as supplementary material and an interactive web interface is available at https://hb.flatironinstitute.org/asdbrowser/.
Code availability
The code used in this study is available from https://hb.flatironinstitute.org/asdbrowser/help.
References
Sanders, S. J. et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature 485, 237–241 (2012).
Iossifov, I. et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature 515, 216–221 (2014).
Yuen, R. K. C. et al. Whole genome sequencing resource identifies 18 new candidate genes for autism spectrum disorder. Nat. Neurosci. 20, 602–611 (2017).
Bernstein, B. E. et al. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Finucane, H. K. et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet. 47, 1228–1235 (2015).
Stenson, P. D. et al. The human gene mutation database: 2008 update. Genome Med. 1, 13 (2009).
Feigin, M. E. et al. Recurrent noncoding regulatory mutations in pancreatic ductal adenocarcinoma. Nat. Genet. 49, 825–833 (2017).
Sanders, S. J. et al. Insights into autism spectrum disorder genomic architecture and biology from 71 risk loci. Neuron 87, 1215–1233 (2015).
Brandler, W. M. et al. Paternally inherited cis-regulatory structural variants are associated with autism. Science 360, 327–331 (2018).
Turner, T. N. et al. Genome sequencing of autism-affected families reveals disruption of putative noncoding regulatory DNA. Am. J. Hum. Genet. 98, 58–74 (2016).
Turner, T. N. et al. Genomic patterns of de novo mutation in simplex autism. Cell 171, 710–722 (2017).
Yuen, R. K. C. et al. Genome-wide characteristics of de novo mutations in autism. NPJ Genom. Med. 1, 16027 (2016).
Yuen, R. K. C. et al. Whole-genome sequencing of quartet families with autism spectrum disorder. Nat. Med. 21, 185–191 (2015).
Michaelson, J. J. et al. Whole-genome sequencing in autism identifies hot spots for de novo germline mutation. Cell 151, 1431–1442 (2012).
Jiang, Y. et al. Detection of clinically relevant genetic variants in autism spectrum disorder by whole-genome sequencing. Am. J. Hum. Genet. 93, 249–263 (2013).
Kong, A. et al. Rate of de novo mutations and the importance of father’s age to disease risk. Nature 488, 471–475 (2012).
Werling, D. M. et al. An analytical framework for whole-genome sequence association studies and its implications for autism spectrum disorder. Nat. Genet. 50, 727–736 (2018).
An, J. Y. et al. Genome-wide de novo risk score implicates promoter variation in autism spectrum disorder. Science 362, eaat6576 (2018).
Lek, M. et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 (2016).
Bernstein, B. E. et al. The NIH Roadmap Epigenomics Mapping Consortium. Nat. Biotechnol. 28, 1045–1048 (2010).
Zhou, J. & Troyanskaya, O. G. Predicting effects of noncoding variants with deep learning-based sequence model. Nat. Methods 12, 931–934 (2015).
Ule, J., Hwang, H.-W. & Darnell, R. B. The future of cross-linking and immunoprecipitation (CLIP). Cold Spring Harb. Perspect. Biol. 10, a032243 (2018).
1000 Genomes Project Consortium. A global reference for human genetic variation. Nature 526, 68–74 (2015).
Kosmicki, J. A. et al. Refining the role of de novo protein-truncating variants in neurodevelopmental disorders by using population reference samples. Nat. Genet. 49, 504–510 (2017).
Aguet, F. et al. Genetic effects on gene expression across human tissues. Nature 550, 204–213 (2017).
Packer, A. Neocortical neurogenesis and the etiology of autism spectrum disorder. Neurosci. Biobehav. Rev. 64, 185–195 (2016).
Krishnan, A. et al. Genome-wide prediction and functional characterization of the genetic basis of autism spectrum disorder. Nat. Neurosci. 19, 1454–1462 (2016).
Greene, C. S. et al. Understanding multicellular function and disease with human tissue-specific networks. Nat. Genet. 47, 569–576 (2015).
Iossifov, I. et al. Low load for disruptive mutations in autism genes and their biased transmission. Proc. Natl Acad. Sci. USA 112, E5600–E5607 (2015).
Valente, E. M. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science 304, 1158–1160 (2004).
Kageyama, R. & Ohtsuka, T. The Notch–Hes pathway in mammalian neural development. Cell Res. 9, 179–188 (1999).
Bertrand, N., Castro, D. S. & Guillemot, F. Proneural genes and the specification of neural cell types. Nat. Rev. Neurosci. 3, 517–530 (2002).
Crosnier, C., Stamataki, D. & Lewis, J. Organizing cell renewal in the intestine: stem cells, signals and combinatorial control. Nat. Rev. Genet. 7, 349–359 (2006).
Eckler, M. J. & Chen, B. Fez family transcription factors: controlling neurogenesis and cell fate in the developing mammalian nervous system. BioEssays 36, 788–797 (2014).
Hormozdiari, F., Penn, O., Borenstein, E. & Eichler, E. E. The discovery of integrated gene networks for autism and related disorders. Genome Res. 25, 142–154 (2015).
Saied-Santiago, K. & Blow, H. E. Diverse roles for glycosaminoglycans in neural patterning. Dev. Dyn. 247, 54–74 (2017).
Chang, W.-H. et al. Smek1/2 is a nuclear chaperone and cofactor for cleaved Wnt receptor Ryk, regulating cortical neurogenesis. Proc. Natl Acad. Sci. USA 114, E10717–E10725 (2017).
Walsh, C. A., Morrow, E. M. & Rubenstein, J. L. R. Autism and brain development. Cell 135, 396–400 (2008).
Weiner, D., Wigdor, E., Ripke, S. & Robinson, E. Polygenic transmission disequilibrium confirms that common and rare variation act additively to create risk for autism spectrum disorders. Nat. Genet. 49, 978–985 (2017).
Liu, Y., Li, B., Tan, R., Zhu, X. & Wang, Y. A gradient-boosting approach for filtering de novo mutations in parent–offspring trios. Bioinformatics 30, 1830–1836 (2014).
Smit, A., Hubley, R. & Green, P. RepeatMasker Open-4.0 (2013).
Moore, M. J. et al. Mapping Argonaute and conventional RNA-binding protein interactions with RNA at single-nucleotide resolution using HITS-CLIP and CIMS analysis. Nat. Protoc. 9, 263–293 (2014).
Darnell, J. C. et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 146, 247–261 (2011).
Deciphering Developmental Disorders Study. Prevalence and architecture of de novo mutations in developmental disorders. Nature 542, 433–438 (2017).
Wright, C. F. et al. Genetic diagnosis of developmental disorders in the DDD study: a scalable analysis of genome-wide research data. Lancet 385, 1305–1314 (2015).
Cotney, J. et al. The autism-associated chromatin modifier CHD8 regulates other autism risk genes during human neurodevelopment. Nat. Commun. 6, 6404 (2015).
Sugathan, A. et al. CHD8 regulates neurodevelopmental pathways associated with autism spectrum disorder in neural progenitors. Proc. Natl Acad. Sci. USA 111, E4468–E4477 (2014).
Harrow, J. et al. GENCODE: the reference human genome annotation for the ENCODE project. Genome Res. 22, 1760–1774 (2012).
Forrest, A. R. R. et al. A promoter-level mammalian expression atlas. Nature 507, 462–470 (2014).
Yan, Q. et al. Systematic discovery of regulated and conserved alternative exons in the mammalian brain reveals NMD modulating chromatin regulators. Proc. Natl Acad. Sci. USA 112, 3445–3450 (2015).
Gene Ontology Consortium. Gene Ontology Consortium: going forward. Nucleic Acids Res. 43, D1049–D1056 (2015).
Mi, H. et al. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 45, D183–D189 (2017).
Geifman, N., Monsonego, A. & Rubin, E. The neural/immune Gene Ontology: clipping the gene ontology for neurological and immunological systems. BMC Bioinformatics 11, 458 (2010).
Van Der Maaten, L. & Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 620, 267–284 (2008).
Acknowledgements
We are grateful to the families participating in the SFARI SSC. This work is supported by NIH grants R01HG005998, U54HL117798 and R01GM071966, HHS grant HHSN272201000054C and Simons Foundation grant 395506 to O.G.T.; NIH grants 1UM1HG008901, NS034389, NS081706 and NS097404 and Simons Foundation grant SFARI 240432 to R.B.D.; and STARR Cancer Consortium Award I10-0056 to C.Y.P. and R.B.D. O.G.T. is a senior fellow of the Genetic Networks program of the Canadian Institute for Advanced Research (CIFAR). R.B.D. is an Investigator of the Howard Hughes Medical Institute. The authors acknowledge all members of the Troyanskaya and Darnell laboratory for helpful discussions. We also thank the SFARI, Simons Foundation and Flatiron Institute, in particular N. Volfovsky and M. Benedetti. We are pleased to acknowledge that a substantial portion of the work in this paper was performed at the TIGRESS high-performance computer center at Princeton University, which is jointly supported by the Princeton Institute for Computational Science and Engineering and the Princeton University Office of Information Technology’s Research Computing department. O.G.T. is a CIFAR fellow.
Author information
Authors and Affiliations
Contributions
J.Z., C.Y.P., C.L.T., R.B.D. and O.G.T. conceived and designed the study. J.Z. and C.Y.P. developed the computational methods and performed the analyses. J.Z. developed the DNA model and C.Y.P. developed the RNA model. C.L.T. designed and performed luciferase assay experiments. Y.Y., C.S., J.J.F. and Y.T. designed and performed the minigene splicing assay and RBP experiments. A.K.W., J.F. and K.Y. developed the web interface. A.P. contributed ideas and insights. J.Z., C.Y.P., C.L.T., R.B.D. and O.G.T. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Note and Supplementary Figures 1–16
Supplementary Table 1
All de novo mutations identified from the WGS cohort with predicted disease impact scores
Supplementary Table 2
Genomic variant set analysis of mutational burden for transcriptional and post-transcriptional disruptions
Supplementary Table 3
NDEA significance levels of proband excess for all genes
Supplementary Table 4
NDEA significance levels of proband excess for all gene sets
Supplementary Table 5
Cluster-specific gene set enrichment for top NDEA significant genes
Supplementary Table 6
Genomic sequences tested in luciferase assays (plasmid backbone pGL4.23)
Supplementary Table 7
List of chromatin profiles used in this study
Supplementary Table 8
List of RBP profiles used in this study
Rights and permissions
About this article
Cite this article
Zhou, J., Park, C.Y., Theesfeld, C.L. et al. Whole-genome deep-learning analysis identifies contribution of noncoding mutations to autism risk. Nat Genet 51, 973–980 (2019). https://doi.org/10.1038/s41588-019-0420-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-019-0420-0
This article is cited by
-
Tapioca: a platform for predicting de novo protein–protein interactions in dynamic contexts
Nature Methods (2024)
-
Quantifying negative selection in human 3ʹ UTRs uncovers constrained targets of RNA-binding proteins
Nature Communications (2024)
-
Fucosyltransferase 8 regulates adult neurogenesis and cognition of mice by modulating the Itga6-PI3K/Akt signaling pathway
Science China Life Sciences (2024)
-
Correcting gradient-based interpretations of deep neural networks for genomics
Genome Biology (2023)
-
DeepASDPred: a CNN-LSTM-based deep learning method for Autism spectrum disorders risk RNA identification
BMC Bioinformatics (2023)