Abstract
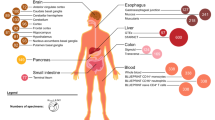
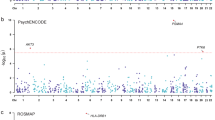
The genetic architecture of psychiatric disorders is characterized by a large number of small-effect variants1 located primarily in non-coding regions, suggesting that the underlying causal effects may influence disease risk by modulating gene expression2,3,4. We provide comprehensive analyses using transcriptome data from an unprecedented collection of tissues to gain pathophysiological insights into the role of the brain, neuroendocrine factors (adrenal gland) and gastrointestinal systems (colon) in psychiatric disorders. In each tissue, we perform PrediXcan analysis and identify trait-associated genes for schizophrenia (n associations = 499; n unique genes = 275), bipolar disorder (n associations = 17; n unique genes = 13), attention deficit hyperactivity disorder (n associations = 19; n unique genes = 12) and broad depression (n associations = 41; n unique genes = 31). Importantly, both PrediXcan and summary-data-based Mendelian randomization/heterogeneity in dependent instruments analyses suggest potentially causal genes in non-brain tissues, showing the utility of these tissues for mapping psychiatric disease genetic predisposition. Our analyses further highlight the importance of joint tissue approaches as 76% of the genes were detected only in difficult-to-acquire tissues.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The protected data for the GTEx project (for example, genotype and RNA-sequence data) are available via access request to dbGaP accession number phs000424.v6.p1. Processed GTEx data (for example, gene expression and eQTLs) are available on the GTEx portal (https://gtexportal.org). The URLs of the summary statistics datasets of all the GWAS meta-analyses analyzed in the paper can be found in Supplementary Table 1.
References
Gratten, J., Wray, N. R., Keller, M. C. & Visscher, P. M. Large-scale genomics unveils the genetic architecture of psychiatric disorders. Nat. Neurosci. 17, 782–790 (2014).
Nicolae, D. L. et al. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 6, e1000888 (2010).
Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427 (2014).
Gamazon, E. R. et al. The convergence of eQTL mapping, heritability estimation and polygenic modeling: emerging spectrum of risk variation in bipolar disorder. Preprint at https://arxiv.org/abs/1303.6227 (2013).
Geschwind, D. H. & Flint, J. Genetics and genomics of psychiatric disease. Science 349, 1489–1494 (2015).
Ozomaro, U., Wahlestedt, C. & Nemeroff, C. B. Personalized medicine in psychiatry: problems and promises. BMC Med. 11, 132 (2013).
Jansen, R. et al. Gene expression in major depressive disorder. Mol. Psychiatry 21, 339–347 (2016).
Bercik, P. et al. The intestinal microbiota affect central levels of brain-derived neurotropic factor and behavior in mice. Gastroenterology 141, 599–609.e3 (2011).
Kato, T. A., Hayakawa, K., Monji, A. & Kanba, S. Missing and possible link between neuroendocrine factors, neuropsychiatric disorders, and microglia. Front. Integr. Neurosci. 7, 53 (2013).
Neale, B. M. et al. Meta-analysis of genome-wide association studies of attention-deficit/hyperactivity disorder. J. Am. Acad. Child Adolesc. Psychiatry 49, 884–897 (2010).
Psychiatric GWAS Consortium Bipolar Disorder Working Group. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat. Genet. 43, 977–983 (2011).
Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium. A mega-analysis of genome-wide association studies for major depressive disorder. Mol. Psychiatry 18, 497–511 (2013).
Pardinas, A. F. et al. Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat. Genet. 50, 381–389 (2018).
Bipolar Disorder and Schizophrenia Working Group of the Psychiatric Genomics Consortium. Genomic dissection of bipolar disorder and schizophrenia, including 28 subphenotypes. Cell 173, 1705–1715.e16 (2018).
Wray, N. R. et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat. Genet. 50, 668–681 (2018).
Howard, D. M. et al. Genome-wide association study of depression phenotypes in UK Biobank identifies variants in excitatory synaptic pathways. Nat. Commun. 9, 1470 (2018).
Demontis, D. et al. Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat. Genet. 51, 63–75 (2019).
Gamazon, E. R. et al. A gene-based association method for mapping traits using reference transcriptome data. Nat. Genet. 47, 1091–1098 (2015).
Huckins, L. M. et al. Gene expression imputation across multiple brain regions provides insights into schizophrenia risk. Nat. Genet. 51, 659–674 (2017).
Foster, J. A. & McVey Neufeld, K. A. Gut-brain axis: how the microbiome influences anxiety and depression. Trends Neurosci. 36, 305–312 (2013).
Leonard, B. E. The concept of depression as a dysfunction of the immune system. Curr. Immunol. Rev. 6, 205–212 (2010).
Sudlow, C. et al. UK Biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 12, e1001779 (2015).
Denny, J. C. et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat. Biotechnol. 31, 1102–1110 (2013).
Xiao, X. et al. Further evidence for the association between LRP8 and schizophrenia. Schizophr Res. https://doi.org/10.1016/j.schres.2017.05.002 (2017).
Storey, J. D. & Tibshirani, R. Statistical significance for genomewide studies. Proc. Natl Acad. Sci. USA 100, 9440–9445 (2003).
The GTEx Consortium. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348, 648–660 (2015).
Wainberg, M. et al. Opportunities and challenges for transcriptome-wide association studies. Nat. Genet. 51, 592–599 (2019).
Qi, T. et al. Identifying gene targets for brain-related traits using transcriptomic and methylomic data from blood. Nat. Commun. 9, 2282 (2018).
Hauberg, M. E. et al. Large-scale identification of common trait and disease variants affecting gene expression. Am. J. Hum. Genet. 101, 157 (2017).
Schizophrenia Psychiatric Genome-Wide Association Study Consortium. Genome-wide association study identifies five new schizophrenia loci. Nat. Genet. 43, 969–976 (2011).
Sekar, A. et al. Schizophrenia risk from complex variation of complement component 4. Nature 530, 177–183 (2016).
Zhang, J. et al. Spatial clustering and common regulatory elements correlate with coordinated gene expression. PLoS Comput. Biol. 15, e1006786 (2019).
Voineagu, I. et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature 474, 380–384 (2011).
Westra, H. J. et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat. Genet. 45, 1238–1243 (2013).
Gamazon, E. R. et al. Using an atlas of gene regulation across 44 human tissues to inform complex disease- and trait-associated variation. Nat. Genet. 50, 956–967 (2018).
Gusev, A. et al. Transcriptome-wide association study of schizophrenia and chromatin activity yields mechanistic disease insights. Nat. Genet. 50, 538–548 (2018).
Jones, A. L., Mowry, B. J., Pender, M. P. & Greer, J. M. Immune dysregulation and self-reactivity in schizophrenia: do some cases of schizophrenia have an autoimmune basis? Immunol. Cell Biol. 83, 9–17 (2005).
Stringer, S., Kahn, R. S., de Witte, L. D., Ophoff, R. A. & Derks, E. M. Genetic liability for schizophrenia predicts risk of immune disorders. Schizophr. Res. 159, 347–352 (2014).
International Consortium for Blood Pressure Genome-Wide Association Studies. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature 478, 103–109 (2011).
Shi, H., Kichaev, G. & Pasaniuc, B. Contrasting the genetic architecture of 30 complex traits from summary association data. Am. J. Hum. Genet. 99, 139–153 (2016).
Gamazon, E. R., Cox, N. J. & Davis, L. K. Structural architecture of SNP effects on complex traits. Am. J. Hum. Genet. 95, 477–489 (2014).
Barbeira, A. et al. Integrating tissue specific mechanisms into GWAS summary results. Preprint at bioRxiv https://doi.org/10.1101/045260 (2017).
Loh, P. R. et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat. Genet. 47, 284–290 (2015).
Chen, G. B. et al. Across-cohort QC analyses of GWAS summary statistics from complex traits. Eur. J. Hum. Genet. 25, 137–146 (2016).
Bulik-Sullivan, B. K. et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47, 291–295 (2015).
The GTEx Consortium. Genetic effects on gene expression across human tissues. Nature 550, 204–213 (2017).
Zhu, Z. et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet. 48, 481–487 (2016).
Shimodaira, H. Approximately unbiased tests of regions using multistep-multiscale bootstrap resampling. Ann. Stat. 32, 2616–2641 (2004).
Yu, G., Wang, L. G., Han, Y. & He, Q. Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012).
Acknowledgements
The GTEx Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health. Additional funds were provided by the National Cancer Institute (NCI), National Human Genome Research Institute (NHGRI), National Heart, Lung, and Blood Institute (NHLBI), National Institute on Drug Abuse (NIDA), National Institute of Mental Health (NIMH) and National Institute of Neurological Disorders and Stroke (NINDS). Donors were enrolled at Biospecimen Source Sites funded by NCI/SAIC-Frederick, Inc. SAIC-F subcontracts to the National Disease Research Interchange (grant no. 10XS170), Roswell Park Cancer Institute (grant no. 10XS171) and Science Care, Inc. (grant no. X10S172). The Laboratory, Data Analysis and Coordinating Center (LDACC) was funded through a contract (grant no. HHSN268201000029C) to The Broad Institute, Inc. Biorepository operations were funded through an SAIC-F subcontract to Van Andel Institute (grant no. 10ST1035). Additional data repository and project management were provided by SAIC-F (grant no. HHSN261200800001E). The Brain Bank was supported by supplements to University of Miami grants (nos. DA006227 and DA033684) and to contract no. N01MH000028. Statistical Methods development grants were made to the University of Geneva (nos. MH090941 and MH101814), the University of Chicago (nos. MH090951, MH090937, MH101820, MH101825), the University of North Carolina at Chapel Hill (nos. MH090936 and MH101819), Harvard University (no. MH090948), Stanford University (no. MH101782), Washington University St Louis (no. MH101810) and the University of Pennsylvania (no. MH101822). E.M.D. is supported by the Foundation Volksbond Rotterdam. E.R.G. is grateful to the President and Fellows of Clare Hall, University of Cambridge, for providing a stimulating intellectual home and benefited from a Fellowship in the College. E.R.G. also acknowledges support from the Academic Medical Center, University of Amsterdam, during the early stages of the study. E.R.G. and N.J.C. acknowledge support from grant no. NIMH R01MH113362. We acknowledge the Psychiatric Genomics Consortium groups for sharing the published meta-analysis data.
Author information
Authors and Affiliations
Contributions
E.R.G. and E.M.D. conceived of and designed the study, analysed the data and drafted the manuscript. E.R.G., A.H.Z. and E.M.D. interpreted the data. N.J.C. and D.D. revised the manuscript critically for important intellectual content. All authors approved of the version of the manuscript to be published.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Gamazon, E.R., Zwinderman, A.H., Cox, N.J. et al. Multi-tissue transcriptome analyses identify genetic mechanisms underlying neuropsychiatric traits. Nat Genet 51, 933–940 (2019). https://doi.org/10.1038/s41588-019-0409-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-019-0409-8
This article is cited by
-
Tissue-specific atlas of trans-models for gene regulation elucidates complex regulation patterns
BMC Genomics (2024)
-
Multiple reaction monitoring assays for large-scale quantitation of proteins from 20 mouse organs and tissues
Communications Biology (2024)
-
Genetic imputation of kidney transcriptome, proteome and multi-omics illuminates new blood pressure and hypertension targets
Nature Communications (2024)
-
The mRNA-lncRNA landscape of multiple tissues uncovers key regulators and molecular pathways that underlie heterosis for feed intake and efficiency in laying chickens
Genetics Selection Evolution (2023)
-
Whole-genome sequencing analysis of suicide deaths integrating brain-regulatory eQTLs data to identify risk loci and genes
Molecular Psychiatry (2023)