Abstract
Allotetraploid cotton is an economically important natural-fiber-producing crop worldwide. After polyploidization, Gossypium hirsutum L. evolved to produce a higher fiber yield and to better survive harsh environments than Gossypium barbadense, which produces superior-quality fibers. The global genetic and molecular bases for these interspecies divergences were unknown. Here we report high-quality de novo–assembled genomes for these two cultivated allotetraploid species with pronounced improvement in repetitive-DNA-enriched centromeric regions. Whole-genome comparative analyses revealed that species-specific alterations in gene expression, structural variations and expanded gene families were responsible for speciation and the evolutionary history of these species. These findings help to elucidate the evolution of cotton genomes and their domestication history. The information generated not only should enable breeders to improve fiber quality and resilience to ever-changing environmental conditions but also can be translated to other crops for better understanding of their domestication history and use in improvement.
Similar content being viewed by others
Main
Each crop has its own unique origin and diversification and domestication histories that have altered its genetic composition, resulting in a variety of physiological and phenotypic differences in comparison to wild relatives1. The genus Gossypium is an attractive model for studying the origin, evolution and domestication of polyploids. It contains two cultivated tetraploids, G. hirsutum L. (AD1) and G. barbadense L. (AD2), which originated in the New World from transoceanic hybridization of an A-genome-like ancestral African species, Gossypium herbaceum or Gossypium arboreum, with a native D-genome-like species, Gossypium raimondii (D5), ~1–1.5 million years ago (MYA)2,3,4. The nascent allotetraploid diverged into five cotton species ~1 MYA (ref. 5). Of these, G. hirsutum and G. barbadense independently evolved and were domesticated in diverse geographic regions. Domesticated G. hirsutum has shown broader adaptability due to its high yield potential with moderate fiber quality and accounts for ~90% of annual world cotton output, whereas G. barbadense produces exceptionally high-quality fibers and confers a competitive advantage to specialty cotton textiles producers.
The global genetic and molecular bases underlying the evolutionary dynamics associated with the origin, speciation and diversification of these two tetraploid species are not well understood. The cultivated tetraploid cottons4,5,6,7 and their corresponding diploid progenitors8,9,10 have been sequenced. However, intergenic DNA such as telomeres, centromeres and repeat-rich regions are often poorly represented in the published draft genomes4,5,6,7, leading to underestimation of many important genomic features found in these regions. Here we report two highly accurate, contiguous, chromosome-scale de novo assemblies of the G. hirsutum and G. barbadense genomes obtained by integrating non-PCR-based short-read sequencing, long-read-based gap closure, scaffolding, and orientation based on 3D proximity information derived from chromosome conformation capture (Hi-C) data and from optical and genetic maps. By using these two well-assembled genomes, we performed genome-wide comparative studies that unraveled the genomic components responsible for contrasting features between the species, demonstrating their origin in homeolog losses, genome restructuring in the postpolyploidization era and altered patterns of gene expression.
Results
Assembly of high-quality genomes
We adopted a hierarchical approach to perform chromosome-scale assembly (Supplementary Fig. 1). For G. barbadense L. cv. Hai7124, about 800 Gb of high-quality sequence (>330× genome coverage) generated from a series of PCR-free libraries, mate-paired libraries and a 10x Genomics library (Supplementary Table 1) was assembled by using the DeNovoMAGIC3 software package (NRGene), which has been used extensively to assemble the complex wheat (Triticum spp.)11,12,13,14, opium15 and maize16 genomes. For Hai7124, we initially generated an assembly that captured 2.23 Gb of sequence (Supplementary Table 2). The initial scaffolds were then corrected and merged by using optical map data (Bionano Genomics) (Supplementary Table 3). Finally, we used 3D proximity information from Hi-C (Supplementary Table 4) and our updated ultra-dense genetic map with 6,165,057 SNPs (Supplementary Table 5) to check, order and orient the resulting superscaffolds. A total of 115 erroneously assembled loci were detected and corrected (Supplementary Table 6). The final Hai7124 (v1.1) assembly captured 2.22 Gb of genome sequence, covering approximately 91.4% of the total genome size, which was estimated to be 2.43 Gb by k-mer analysis (Supplementary Fig. 2). The contiguity of the newly assembled genome was 47- and 90-fold greater than that of the two published draft genomes of G. barbadense (scaffold N50 = 23.44 Mb versus 0.50 Mb (ref. 5) and 0.26 Mb (ref. 7)). Approximately 98.2% of the 2.22-Gb assembled genome was assigned to 26 chromosomes (Table 1 and Supplementary Table 7). The gap size (Ns) was substantially reduced to 32.46 Mb in Hai7124 (Supplementary Fig. 3).
We also generated an improved de novo–assembled genome for G. hirsutum L. acc. TM-1 (Supplementary Tables 8 and 9). The total assembly size was 2.30 Gb, with the longest scaffold 41.92 Mb in length. About 97.4% of the 2.30-Gb assembled genome was oriented and organized into 26 chromosomes (Table 1 and Supplementary Table 10). In comparison with two recently published genome assemblies for this accession, the updated TM-1 reference assembly (v2.1) showed higher contiguity, that is, ~10- to 20-fold longer scaffold N50 values (15.5 Mb versus 1.6 Mb (ref. 4) and 0.764 Mb (ref. 6)). The total size of the oriented assembled genome without connective N sequences was dramatically increased (2,211 Mb for TM-1 (v2.1) versus 1,667 Mb for TM-1 (v1.1)4) (Supplementary Fig. 4). We validated the contiguity and accuracy of the new assembly by using the same 36 BAC sequences (Supplementary Fig. 5 and Supplementary Table 11) and integrated genetic and cytogenetic maps of the A12 and D12 homeologous chromosomes from FISH (Supplementary Fig. 6) as we reported previously4.
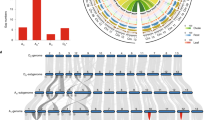
We successfully assembled and filled the gaps in the new genomes, and erroneous assemblies, especially those spanning centromeric regions, were corrected (Fig. 1a,b and Supplementary Figs. 7 and 8). The alignment between Hai7124 (v1.1) and G.barbadense accession 3-79 was largely incomplete (Fig. 1c and Supplementary Fig. 8), probably owing to the fragmented nature of the 3-79 assembly (scaffold N50 = 260 kb, scaffold number = 29,751)7. The highly consistent alignment of the genome sequences to the corresponding optical maps (88% mapping rate in TM-1 and 91% in Hai7124) (Fig.1a,b and Supplementary Figs. 7 and 8) further validates the high contiguity and accuracy of our new genome assemblies.
a, Collinearity on chromosome A01 for the TM-1 (v1.1) and TM-1 (v2.1) genomes. The red region in the blue bar indicates the centromeric region identified by alignment of CRGs. Gray lines indicate collinearity between the two genomes; each line represents a unique mapping alignment >5 kb in length. Yellow bars show the CenH3-binding regions in each genome; adjusted ChIP-seq reads were calculated for a 100-kb sliding window at 10-kb intervals. GC content, repeat distribution and gene density in a 1-Mb sliding window are shown at 100-kb intervals. A comparison of TM-1 Bionano optical maps (blue) and the TM-1 (v2.1) genome (green) is shown. Right, TM-1 Hi-C contact data mapped onto the TM-1 (v2.1) genome. The heat map represents the normalized contact matrix. b, Collinearity on chromosome A01 for the Hai7124 (v1.1) and 3-79 genomes. Data are presented as in a. c, Analysis of the synteny among multiple Gossypium genomes. Macrosynteny connecting blocks of >30 one-to-one gene pairs is shown. Blue links indicate translocation between the D subgenome of allotetraploid cotton and the D genome of diploid cotton; yellow links indicate translocation between the A genome and the D genome in diploid cotton; and green links indicate translocation between the A subgenome of allotetraploid cotton and the A genome of diploid cotton. d, MCScanX was performed to identify the collinearity between genes in the TM-1 (v2.1) and Hai7124 (v1.1) genomes. Each block includes >30 gene pairs.
Notable improvement in the assembly of centromeres
Fine-scale genetic and physical mapping of centromeres in multiple crop species remains a mammoth task. In total, 955, 1,012, 205, 284 and 100 CRG (centromere retroelement Gossypium)17 homologs were identified in the centromeric region of each chromosome for TM-1 (v2.1), Hai7124 (v1.1), TM-1 (v1.1)4, Xinhai 21 (ref. 5) and 3-79 (ref. 7), respectively, by using our previously described approach4. The average length of the centromeric region predicted for each chromosome with a 95% confidence interval around the median was 270 kb in Hai7124 (v1.1) and 358 kb in TM-1 (v2.1), which is substantially less than the 9.23 Mb in TM-1 (v1.1)4, the 6.80 Mb in Xinhai 21 (ref. 5) and the 11.81 Mb in 3-79 (ref. 7) (Supplementary Fig. 9 and Supplementary Tables 12–16).
The centromeric DNA regions associated with centromere-specific H3 (CenH3)18 were further identified by ChIP-seq using an antibody to cotton CenH3. In total, 66.72 and 50.16 million reads (151 bp) were obtained from the TM-1 and Hai7124 ChIP-seq libraries, respectively. In FISH experiments, distinct and high-intensity signals were observed in the centromeric region of each chromosome, further confirming the CenH3 ChIP DNA enrichment (Supplementary Fig. 10). Approximately 22.10 and 17.04 million sequence reads were mapped to unique sites in the genomes for TM-1 (v2.1) and Hai7124 (v1.1), respectively. As a result, potential centromeric regions from all 26 chromosomes of the Hai7124 (v1.1) assembly and 24 chromosomes of the TM-1 (v2.1) assembly (with the exception of D02 and D08) were identified (Fig. 1a,b, Supplementary Figs. 7 and 8, and Supplementary Table 17). The CenH3 ChIP-seq data mapped to a sharp interval on each chromosome, ranging from 0.60 to 2.85 Mb in length, which was highly overlapped by the region identified by the CRGs (Supplementary Tables 12 and 13). However, the CenH3 ChIP-seq reads could not be mapped onto the TM-1 (v1.1) assembly (Fig.1a and Supplementary Figs. 7 and 8), likely owing to the fragmented nature of its centromeric regions. Moreover, centromeres showed consistent localization along the same chromosome in the two cotton species (Supplementary Table 17), implying a high level of genome conservation and a relatively short time to divergence. In plants, extensive studies have been conducted to isolate and map the DNA corresponding to CenH3-associated centromeric chromatin by using ChIP-seq19,20,21. However, poorly assembled reference centromeres often hamper the deployment of ChIP-seq, which has resulted in poor characterization of the centromeres of the sequenced plants22,23,24. Until now, only five centromeres from Brachypodium distachyon, a small-genome grass, had been fully sequenced and assembled, which was possible because these centromeres have a relatively low percentage of highly repetitive DNA (~21.4%)25,26. These results demonstrate that our new genome assemblies have made substantial improvement in sequence continuity in almost all regions, including repetitive-DNA-rich centromeric regions.
Gene prediction and annotation
A total of 72,761 and 75,071 high-confidence protein-coding genes (PCGs) were predicted for TM-1 (v2.1) and Hai7124 (v1.1), respectively, of which >96% were supported by full-length transcript data (Supplementary Tables 18 and 19). For most of these genes (95%; n = 100), there was complete accuracy between the identified exon–intron boundaries and those determined by using mapped RNA-seq reads (Supplementary Fig. 11 and Supplementary Table 20). Benchmarking Universal Single-Copy Orthologs (BUSCO) analysis27 revealed full gene content for 97.5% of PCGs in TM-1 and for 97.3% of PCGs in Hai7124 (Table 1 and Supplementary Table 21); of these genes, the majority were retained in duplicate, largely owing to the allotetraploid nature of the genomes. In total, 5,558 and 5,606 genes in TM-1 and Hai7124, respectively, were putative transcription factors belonging to 58 families, representing 7.6% and 7.5% of the PCGs (Supplementary Table 22).
A total of 1,460.46 Mb and 1,374.61 Mb of transposable element (TE) sequence accounted for 62.2–63.9% of the total genome size for TM-1 and Hai7124, respectively (Supplementary Tables 23 and 24). The TE content in the A subgenome (979.05 Mb in TM-1 and 922.04 Mb in Hai7124) was twice that identified in the D subgenome (443.23 Mb in TM-1 and 420.66 Mb in Hai7124) (Supplementary Tables 23 and 24). The majority of TEs were Gypsy long terminal repeat (LTR) retroelements, including 41.4% of TEs in G. hirsutum and 39.5% of TEs in G. barbadense. The distribution of the divergence rate for each type of TE was similar for the two genomes (Supplementary Fig. 12). Furthermore, we predicted the insertion time of LTRs by examining the sequence divergence at both ends of intact LTRs between TM-1 and Hai7124. Two expansions were identified that were distinct from the amplification wave of complete LTR elements (Supplementary Fig. 13). These two recent LTR insertion events seem to have occurred around 2 MYA and 13 MYA, close to the time of tetraploid cotton formation (1–2 MYA)4,10 and the divergence time between diploid A- and D-genome species (2–13 MYA)6.
Speciation and genome evolution
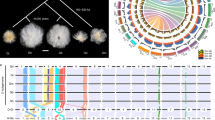
We resequenced 17 accessions representing the D genome and wild AD genome (Supplementary Table 25), finding that G. raimondii (D5) was the closest relative to the D subgenome for all five allotetraploids (Supplementary Fig. 14). Furthermore, whole-genome sequence alignment of our current reference-quality assemblies to genomes for the diploid progenitors G. arboreum (A genome)8 and G. raimondii (D genome)10 demonstrated that the two tetraploids have highly collinear relationships with their diploid progenitors. However, some reciprocal translocations were observed along chromosomes 1–3, as well as along chromosomes 4 and 5 (Fig. 1c and Supplementary Table 26), in accordance with earlier reports28,29,30,31. A total of 2.2 Gb of sequence from Hai7124 (v1.1) could be mapped onto the TM-1 (v2.1) genome, suggesting that the collinearity and gene order for these species are largely conserved (Fig. 1d). These data support the idea that G. hirsutum and G. barbadense originated from a common allotetraploid ancestor.
The nonsynonymous (Ka) and synonymous (Ks) substitution rates for 22,054 orthologous gene sets were compared between G. arboreum8, G. raimondii10, and the A and D subgenomes of the tetraploid species. On the basis of this, the divergence time for the A and D progenitor genomes was estimated to be around 6.2–7.1 MYA, and the allotetraploid formed around 1.7–1.9 MYA (Fig. 2a and Supplementary Table 27). The divergence of G. barbadense and G. hirsutum occurred ~0.4–0.6 MYA (Ks peaks at 0.002 and 0.003, respectively).
a, Evolutionary analysis of the G. barbadense and G. hirsutum genomes. The Ks distribution is shown for orthologs in the cotton genomes; data are grouped into Ks units of 0.001. Species are represented by their initials; At, A subgenome; Dt, D subgenome. b, Comparison of the codon substitution rate distribution between the two subgenomes of TM-1 and Hai7124 and their progenitors. The central line for each box plot indicates the median, the top and bottom edges indicate the seventy-fifth and twenty-fifth percentiles, and the whiskers extend to 1.5 times the interquartile range. Asterisks indicate significant differences (Wilcoxon rank-sum test, P < 0.05). c, Distribution of SNPs, indels, PAVs, inversions and translocations across the TM-1 genome in comparison to the Hai7124 genome. Regions of >500 kb that showed lower than expected SNP frequencies are indicated by green blocks along the bottom track, and those with higher than expected SNP frequencies are indicated by red blocks.
We identified 9,451 one-to-one orthologous gene sets among the genome assemblies for Theobroma cacao32, the A and D diploid progenitors, and the A and D subgenomes of our two currently assembled cultivated species, which suggests an accelerated evolution rate in allotetraploids in comparison to their diploid progenitors. A faster evolution rate was observed in allotetraploid cottons in comparison to their living diploid progenitors; in the A subgenome in comparison to the D subgenome; and in G. barbadense in comparison to G. hirsutum (Fig. 2b, Supplementary Fig. 15 and Supplementary Table 28). In addition, greater numbers of lost, disrupted and positively selected genes were found in the A subgenome (Supplementary Fig. 16), indicating that the A subgenome might have experienced strong selection pressures during evolution.
Deep sequencing data (average coverage of 60×) from nine G. barbadense and ten G. hirsutum accessions (Supplementary Table 29) showed high divergence between G. hirsutum and G. barbadense, as well as between the wild G. hirsutum race yucatanense and the domesticated and/or improved accessions for G. hirsutum (Supplementary Fig. 17a). Furthermore, a long reciprocally introgressed region exhibiting extremely low levels of polymorphism, extending from 43.10 Mb to 92.00 Mb on chromosome A01, was found by comparing the G. hirsutum race with G. barbadense (Supplementary Fig. 17b), supporting the idea that hybridization occurred between domesticated species and their wild ancestors in the spread of domestication and/or diversification, as we reported previously33. In addition, Xinhai 25 showed introgression from the G. hirsutum race at 48.70–57.90 Mb and 81.60–92.00 Mb. Junhai 1 showed introgression from the G. hirsutum race at 48.40–54.50 Mb and 75.00–92.00 Mb (Supplementary Fig. 17b and Supplementary Table 30). The introgressed region from the long-lived, perennial G. hirsutum race on chromosome A01 was found in nine G. barbadense accessions collected from Egyptian, American Pima and Central Asian ELS (extra-long-staple) cottons, but not in the Tanguis landrace, which produces coarse fibers with medium staple length, which is typical of current cottons from Peru. It was confirmed that G. barbadense originated in a region spanning northwestern Peru and southwestern Ecuador34. Introgression of wild and/or domesticated alleles from G. hirsutum may have induced changes in ‘morpho-physiological traits’, helping in the domestication of Sea Island cotton (G. barbadense). This analysis of introgressed regions further revealed that there were two genotypes among the Central Asian ELS cottons: one was the same as the Egyptian or American Pima type, represented by Hai7124 and Xinhai 21, and the other is a new type, represented by Junhai 1 and Xinhai 25, selected from cultivar 9122 from the former Soviet Union. Therefore, the spread and adaptation of domesticated cottons to different agro-ecological and cultural environments have led to phenotypic and genetic divergence among domesticated populations to produce two cultivated allotetraploid cotton types.
Postpolyploidization genome restructuring
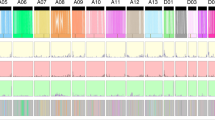
Genetically, G. hirsutum and G. barbadense are cross-compatible, but their interspecific hybrids exhibit genetic breakdown during segregation, which needs to be investigated. Alignment of Hai7124 to the TM-1 genome identified 13,845,619 SNPs and 1,358,773 indels (Supplementary Table 31), with 5.98 SNPs and 0.59 indels per kilobase on average. We also detected 34 SNP-poor and 5 SNP-rich regions, covering 90.4 Mb and 6.2 Mb of the genome, respectively (Fig. 2c and Supplementary Table 32). Of these variations, 200,528–213,634 SNPs and 7,926–8,453 indels were found in coding regions. A total of 10,366 genes with sequence variations (GSVs) resulting in gain or loss of stop codons and frameshifts, among other changes, were identified (Supplementary Table 33). These GSVs were classified into three types: type I consisted of 3,029 GSVs identified in Hai7124 but annotated only in TM-1 and type III consisted of 3,556 GSVs identified in TM-1 but annotated only in Hai7124. The 3,781 type II GSVs were annotated in both species (Fig. 3a). 69.7% of the species-specific GSVs were completely validated in the 19 deep-sequenced accessions (Supplementary Tables 34 and 35). Gene Ontology (GO) enrichment analysis showed that the type I GSVs were enriched for genes involved in defense response and DNA integration, the type II GSVs were enriched for genes involved in response to stress and oxidation–reduction processes, and the type III GSVs were enriched for genes involved in DNA integration and cell recognition. The variations mapping to these GSVs may be among the causative mutations that have led to the phenotypic divergence between these two species. For instance, among the type II GSVs, a 2-bp (CA) deletion in WLIM1a was identified in Hai7124 (GB_D06G0243) that resulted in premature termination of translation. Thus, in comparison to the wild-type homolog in TM-1 (GH_D06G0228), there were 23 amino acids deleted from the encoded protein in Hai7124, including 5 in the second LIN-11, Isl1 and MEC-3 (LIM) motif (Supplementary Fig. 18a). This species-specific indel was further confirmed in all 19 accessions (Supplementary Fig. 18b). The protein encoded by WLIM1a has a role in cotton fiber elongation as well as secondary cell wall (SCW) formation35.
a, Classification of GSVs identified through comparison of TM-1 and Hai7124. Type I and type III GSVs correspond to genes annotated in only TM-1 and Hai7124, respectively. Type II GSVs correspond to genes containing sequence variations between TM-1 and Hai7124. The y axis shows the length ratio of GSVs between TM-1 and Hai7124. b, Length and distribution of TE insertions and deletions identified through comparison of TM-1 and Hai7124. c, TE annotation in the peaks within the regions at 3 kb, 4.9 kb and 11 kb highlighted in b.
By comparing TM-1 and Hai7124, we further identified 10,377 presence regions (101 bp to 99,272 bp) and 12,903 absence regions (101 bp to 96,795 bp) (Fig. 3b), 10 inversions (1,111 bp to 44,679 bp) and 32 translocations (2,530 bp to 11,439 bp) (Supplementary Table 36). Among these, 1,358 structural variations were in genes (Supplementary Table 37), and 70.1% of these were consistent with the classification of the 19 deep-sequenced accessions (Supplementary Table 38). The presence/absence variants (PAVs) were distributed unevenly across the genomes; the majority of these were Copia elements (LTR type) found in coding DNA sequence (Supplementary Fig. 19a,b). The expression level of genes with LTR insertions was lower than that of genes in which PAVs occurred in upstream or downstream sequences or in introns and that of all PCGs expressed in root, stem, leaf or fiber (Supplementary Fig. 19c), but these genes showed much more differential expression between G. hirsutum and G. barbadense (Supplementary Fig. 19d). Of the three major PAV types, the first (74%; ~3 kb) and second (94%; ~11 kb) types were caused by Gypsy LTR retroelements, whereas the third type (68%; ~4.9 kb) was mostly caused by Copia LTR retroelements (Fig. 3c). These structural variations were found to be associated with loss of function of 820 and 791 genes in TM-1 and Hai7124, respectively (Supplementary Table 39). In G. barbadense, we detected a PAV in an α-expansin gene with fiber-specific expression (GB_A10G1730; deletion of 450 bp within exon 3), which resulted in a truncated protein lacking the C-terminal polysaccharide-binding domain as compared with the G. hirsutum homolog (GH_A10G1626, also known as GhEXP1). The protein encoded by this truncated ELS-fiber-related gene can significantly enhance fiber length, fineness and strength through cell wall restructuring in G. hirsutum36. This species-specific deletion in the expansin gene (GB_A10G1730 versus GH_A10G1626) was found in nine of the G. barbadense accessions examined. Furthermore, within the SCW-biosynthesis-related cellulose synthase CESA 7 clade (Supplementary Fig. 20a), a large TE insertion (9,427 bp) in GH_A07G0437 resulted in the absence of 53 amino acids from all ten G. hirsutum accessions but not the nine G. barbadense lines except for Pima S-4 (Supplementary Fig. 20b). The corresponding wild-type CESA 7 gene (GB_A07G0431) showed high expression at 20 days post anthesis (DPA) during the SCW biosynthesis stage in all three G. barbadense accessions examined, suggesting that it potentially has a role in the biosynthesis of more cellulose to develop ELS fibers (Supplementary Fig. 20c). These TE insertions might also have had a key role in cotton domestication and diversification.
Moreover, we identified 3,905 copy number variations (CNVs) of sequences that were either gained or lost in the 19 cotton accessions in comparison to TM-1 (Supplementary Table 40). The genes within CNVs were significantly enriched for genes involved in defense response (P = 2.60 × 10–12, Fisher’s exact test), suggesting that they have a role in adaptation to various environments.
Genomic insights into development of extra-long fibers in G. barbadense
We measured dynamic changes in the elongating fibers of TM-1 and Hai7124 plants from 5 to 40 DPA and observed a nearly linear increase in fiber length at 5–15 DPA in TM-1 plants, whereas this increase occurred at 5–20 DPA in Hai7124 plants (Fig. 4a). The maximum elongation rate was achieved at 15–20 DPA; this rate suddenly decreased at 25 DPA in TM-1 fibers but was retained until 30 DPA in Hai7124 fibers (Fig. 4b). The extended elongation phase in Hai7124 plants (5–30 DPA) most likely leads to development of longer fibers in G. barbadense as compared to TM-1 plants (elongation from 5–25 DPA).
a, Phenotypes of fiber-bearing seeds in Hai7124 and TM-1 plants. Numbers above the graphs are DPA. Scale bar, 10 mm. b, Fiber elongation patterns for TM-1 and Hai7124. Each value is the mean ± s.d. of fiber length for at least ten seeds from three individual plants at a given time point. c–e, Comparison of the content of the main osmotically active solutes soluble sugar (c), malate (d) and K+ (e) between TM-1 and Hai7124 fibers. Mean values and s.e.m. were calculated from three biological replicates for soluble sugar and from six biological replicates for K+ and malate. DW, dry weight. f, Transmission electron microscopy (TEM) analysis of epidermal cell structure in 5-DPA ovule, showing the large central vacuole in the fiber cell compared with fiber base and epidermal cell.Scale bars, 10 µm in the figure and 100 nm in the inset. PD, plasmodesmata; C, cytosol; V, vacuole; CW, cell wall. g, Model depicting the roles of open PD and the genes related to the main osmotically active solutes in fiber elongation. Suc, sucrose; M, malate; Fru, fructose; Glu, glucose. h–k, qRT–PCR analysis of TST1 (h), VIN1 (i), ALMT16 (j) and NHX1 (k) in fibers from TM-1 plants at 5–25 DPA and from Hai7124 plants at 5–30 DPA. Three biological replicates were assessed per reaction, each with two technical replicates (using the same sample). Each value represents the mean ± s.e.m. **P < 0.01, Student’s t test. The expression of TST1, VIN1, NHX1 and ALMT16 in fibers from TM-1 plants at 30 DPA is not shown because RNA could not be extracted from fibers at this time point.
Fiber elongation is driven by cell turgor created by the influx of sucrose and potassium ions (K+) and local production of malate together with high expression of expansion-related genes encoding proteins that loosen the cell wall matrix37,38. The content of the osmotically active solutes in elongating fibers was substantially increased in Hai7124 plants from 10 to 15 DPA, and this high level was sustained from 15 DPA onward (Fig. 4c–e). A large central vacuole occupied 98% of the volume of quickly elongating fiber cells (Fig. 4f); such vacuoles store the main osmotically active solutes that impose maximal turgor pressure for fiber elongation .
To the extent of our knowledge, the underlying molecular basis of ELS fiber production has not previously been investigated. Here we found 45,129 and 45,328 genes expressed (FPKM > 1) in developing single-celled fibers of TM-1 and Hai7124 plants, respectively. Kyoto Encyclopedia of Genes and Genomes (KEGG) annotation of these genes revealed that genes associated with membrane transport, transcription, and glycan biosynthesis and carbon metabolism were significantly enriched for expression in Hai7124 as compared to TM-1 fibers (Supplementary Fig. 21a), highlighting the species-specific role of these genes in fiber elongation. Furthermore, the sucrose transporter (GbTST1), Na+/H+ antiporter (GbNHX1) and aluminum-activated malate transporter (GbALMT16) genes whose expression was detected in the tonoplast of Hai7124 fibers had a longer period of expression than the corresponding genes in TM-1 (Fig. 4g–k and Supplementary Table 41). These transporters pump more sucrose, K+ and malate into the vacuole39,40,41,42,43,44,45. Moreover, sucrose is further degraded into hexose by the vacuole-localized vacuolar invertase (VIN1) to enhance the osmotic potential required for fiber elongation46,47. Interestingly, GbVIN1 also showed prolonged expression in Hai7124 as compared to TM-1 fibers (Fig. 4i).
Unlike malate, which is synthesized in the cytosol of fibers, sucrose and K+ enter fiber cells through the plasma membrane or plasmodesmata (PD)47,48. In Hai7124 plants, the PD remained open for a longer time (5–15 DPA) to import sucrose into the cytosol of developing fiber cells from underlying seed coat cells than it did in TM-1 plants (5–10 DPA) (Supplementary Fig. 21b). Conversely, expression of the plasma membrane–localized SUTs and SWEETs responsible for sucrose transport is almost completely silenced during this stage49. All these findings demonstrate that expression of GbVIN1, GbTST1, GbNHX1 and GbALMT16 and PD opening for a relatively extended time period could be responsible for the accumulation of more soluble sugar, K+ and malate, thereby leading to production of longer fibers in ELS cotton.
In total, there were 1,464 genes representing expansions in 446 gene families (Supplementary Table 42) in the Hai7124 genome, significantly more than the 696 genes found in the TM-1 genome (P = 6.27 × 10–60, Fisher’s exact test) (Supplementary Table 43). For instance, the 12 genes in class I of the ADP-ribosylation factor (ARF) GTPase family, forming core components of vesicle transport machinery and determining epidermal cell polarity50,51, correspond to a specific expansion in Hai7124 (Supplementary Fig. 22a). These ARF GTPase genes were predominantly expressed during fiber elongation (10 DPA) and SCW thickening (20 and 25 DPA), indicating that they potentially have a role in the development of longer and stronger fibers in ELS cotton (Supplementary Fig. 22b).
Divergent evolution of genes involved in abiotic stress tolerance
No significant differences in response to salinity or drought stresses were observed between TM-1 and Hai7124 (Supplementary Fig. 23a,b). However, TM-1 seedlings were more tolerant to heat and cold stresses than Hai7124 seedlings (Fig. 5a–d), suggesting that selection after domestication was aimed at developing the high yield and adaptation abilities of G. hirsutum. We detected 16,029 versus 5,270 genes conferring resistance to heat stress (P = 0, Fisher’s exact test) and 8,725 versus 12,475 genes conferring resistance to cold stress (P = 5.079 × 10–142, Fisher’s exact test) in TM-1 and Hai7124, respectively (Supplementary Table 44). In total, 51 genes including PIPK, PIP (plasma membrane–intrinsic protein), CaM genes and downstream transcriptional regulator (HSF) genes involved in the ability of plants to sense heat52 showed differential responses to heat stress between TM-1 and Hai7124 (Supplementary Tables 41 and 45). Most of these (39 genes), especially the CaM genes, were specifically up- or downregulated under heat stress in G. hirsutum (Fig. 5b). The ethylene signaling pathway was also activated under heat stress in TM-1, as nine each of the genes encoding ethylene receptors and ethylene response factors (ERFs) were dramatically upregulated in TM-1 (Fig. 5b,c, Supplementary Fig. 23c and Supplementary Tables 41 and 45), elucidating their role in regulating EIN3 and EIL1 activation under heat stress53.
a,d, Phenotypic comparison of TM-1 and Hai7124 cotton seedlings in response to heat (a) and cold (d) stress. Photographs were taken under normal conditions or for seedlings subjected to heat treatment at 37 °C or cold treatment at 4 °C for 120 h followed by recovery for 24 h. Scale bars, 10 cm. b,e, Heat maps showing differentially expressed genes involved in the ethylene signaling pathway for heat stress (b) and the ABA signaling pathway for cold stress (e) in TM-1 and Hai7124. Red, upregulation; yellow, downregulation; blue, no difference in expression; white, absence of an orthologous gene. c, A proposed model showing that the ethylene and plasma membrane signaling pathways cooperatively enhance heat tolerance in G. hirsutum. These pathways work together to maintain high transcriptional activities under heat stress, whereas no difference in expression is observed in G. barbadense. f, Diagram of the cold-responsive transcriptional network in cotton. The ABA and calcium signaling pathways were activated in G. hirsutum, whereas only a small portion of the differentially expressed genes were activated in G. barbadense. Upward- and downward-pointing arrows indicate up- and downregulated genes; red and blue numbers correspond to the number of differently expressed genes in TM-1 and Hai7124, respectively.
TM-1 seedlings also showed greater tolerance of cold stress than Hai7124 seedlings (Fig. 5d). In total, 23 genes from the abscisic acid (ABA) signaling pathway that encode ABA receptors (PYR/PYL), the SnRK protein kinase and the PP2C protein phosphatase kinase (ABI/AHG/PP2C)54 showed differential responses to cold in TM-1 and Hai7124 (Fig. 5e,f). Most genes, especially those encoding the ABA receptors ABAR (CHLH), PYR1, PYL3, PYL8 and PYL11, were specifically upregulated under cold stress in TM-1 (Supplementary Table 46), and this upregulation was confirmed by qRT–PCR analysis (Supplementary Fig. 23d). These genes have in part facilitated the adaptation of Upland cotton to new environments.
Discussion
Here we performed de novo assembly of the genomes of two cultivated allotetraploid cotton species by integrating data from Illumina PCR-free short-read sequencing, 10x Genomics sequencing, Hi-C, and optical and super-dense genetic maps. These efforts resulted in substantial improvements to the contiguity and accuracy of assembly, with a notable improvement in the assembly of centromeres. By analyzing the two resulting high-quality genome assemblies, we determined that tissue- and/or developmental-stage-specific expression and divergent neo- and subfunctionalization of homeologs between G. hirsutum and G. barbadense may have led to the variable adaptation responses of these two species after the formation of allotetraploid cottons. Cotton evolutionary processes are also partially driven by the cultural and economic importance of cotton to humans. To our knowledge, this is the first comparison of these two important allotetraploid cotton species over the entire genome. These results should improve understanding of crop evolution, domestication and diversification and should lead to the discovery of novel domestication-related genes conferring agronomically beneficial traits in future breeding programs and evolutionary trajectories in plants.
Methods
DNA extraction and sequencing
Genomic DNA from G. hirsutum L. acc. TM-1 (ref. 55) (genetic standard) and G. barbadense L. cv. Hai7124 (resistant to verticillium wilt) seedlings was isolated by a modified CTAB method56. Five size-selected DNA libraries (470 bp to 10 kb) and three mate-pair libraries (2–5, 5–7 and 7–10 kb) were constructed. To limit sequencing biases across genomic regions with varying GC content, libraries with insert sizes of ~470 and 800 bp were constructed by using TruSeq DNA Sample Preparation Kit version 2 with no PCR amplification. The library with an insert size of ~470 bp was sequenced on a HiSeq 2500 v2 instrument in rapid mode as 2 × 265 bp reads, which produced ‘stitched’ reads of ~265–520 bp in length. The 800-bp shotgun library and the mate-pair libraries were sequenced on a HiSeq 4000 instrument as 2 × 150 bp reads. Construction and sequencing of paired-end and mate-pair libraries were carried out at the Roy J. Carver Biotechnology Center at the University of Illinois at Urbana-Champaign.
DNA molecules longer than 50 kb prepared with the nuclei method57 were used to construct a 10x Genomics library that was sequenced on the GemCode platform (10x Genomics, Pleasanton) at the Hudson Alpha Institute for Biotechnology. About 800 Gb of sequencing data was produced for both TM-1 and Hai7124, respectively (Supplementary Table 1).
Initial genome assembly by DeNovoMAGIC3
A flowchart of contig, scaffold and chromosome assembly in this study is shown in Supplementary Fig. 1. Initial genome assembly was performed by deploying the DeNovoMAGIC3 software platform (NRGene), a de Bruijn graph–based assembler. This task was accomplished by using accurate read-based traveling in the graph that iteratively connected consecutive contigs over local repeats to generate long scaffolds. An additional long-range barcoded DNA library (Chromium system by 10x Genomics) was sequenced to support haplotyping of heterozygous genomes, scaffold validation and further longer-range extension of the scaffolds. The algorithm has been described in detail in wheat D-genome assembly11. de Bruijn graphs (k-mer = 127 bp) of contigs were built from all paired-end and mate-pair reads. Next, paired-end reads were used to find reliable paths in the graph between contigs to resolve repeats and extend contigs.
The barcoded reads from the 10x Genomics library were mapped to the assembled scaffolds, and clusters of reads with the same barcode mapped to adjacent contigs in the scaffolds were identified as being part of a single long molecule. Each scaffold was scanned with a 20-kb window to ensure that the number of distinct clusters that covered the entire window (indicating support for this 20-kb connection by several long molecules) was statistically significant with respect to the number of clusters that spanned the left and right edges of the window. If a potential scaffold assembly error was detected, the scaffold was broken at the two edges of the suspicious 20-kb window. Finally, the barcodes mapping to the scaffold edges were compared (first and last 20-kb sequences) to generate a scaffold graph with a link connecting two scaffolds with more than two shared barcodes. Linear scaffold paths in the scaffold graph were composed into the final scaffold output of assembly. Initial genome assemblies are summarized in Supplementary Tables 2 and 8.
Integrating Bionano optical maps with the initial assembly
High-molecular-weight DNA was extracted from yellowish cotyledons with a Bionano Plant Tissue DNA isolation kit (Bionano Genomics) and digested with Nt.BssSI (New England Biolabs). After labeling and staining, DNA was loaded onto the Saphyr chip for sequencing. 457 and 449 Gb of data (>150 kb) for TM-1 and Hai7124, respectively, were collected and converted into a BNX file by AutoDetect software to obtain basic labeling and DNA length information. The filtered raw DNA molecules in BNX format were aligned, clustered and assembled into the BNG map by using the Bionano Genomics assembly pipeline. Alignment of sequence assemblies with the BNG map was computed with RefAligner, and visualization of alignment was performed with snapshot in IrysView. Combination of the genome maps with the initial assembly to produce a hybrid scaffold was performed sequentially with the two genome maps (Supplementary Table 3).
Construction of the updated POPSEQ genetic map
We constructed an augmented POPSEQ genetic map by sequencing 241 F2 individuals (approximately 5× genome coverage) derived from a cross between TM-1 and Hai7124, including 59 individuals used to construct the previous genetic map58. After SNP calling and genotyping of the 241 F2 individuals, simple SNPs from the parents and SNPs with the same genotype in the population were classified into a bin. Bins were assigned to linkage groups by JoinMap 3.059 with a minimum logarithm of odds (LOD) score of 13 and ordered by MSTmap60.
Hi-C library construction and data collection
We created three Hi-C libraries for TM-1 and Hai7124 by using the method described previously61. Libraries were subjected to sequencing on the Illumina HiSeq X Ten platform. Information about raw data is given in Supplementary Table 4.
Ordering and orientation of the Bionano-assembled scaffolds by Hi-C and POPSEQ
BNG maps were ordered and oriented on the basis of the POPSEQ genetic map. Hi-C data were then used to correct the Bionano assemblies. Hi-C reads were mapped to the assembled scaffolds with BWA62 (version 0.7.15-r1140). PCR duplicates were removed with Novosort v1.04.06 (http://www.novocraft.com/), and reads were excluded from subsequent analysis if they did not align within 500 bp of a restriction site or did not uniquely map. The initial ordered scaffolds were used to make the Hi-C map, and the scaffolds were then divided into bins of 500 kb in size and –log10 (number of Hi-C links) was calculated for all bins. The initial order and orientation were corrected and verified manually by using the Hi-C linking information; heat maps of the Hi-C data were plotted with HiCPlotter63.
Assessment of TM-1 assembly using 36 completely sequenced BACs
The 36 full-length BAC sequences64 were split into 1-kb continuous windows and aligned to the TM-1 (v2.1) assembly by using BWA62 (version 0.7.15-r1140) with the MEM algorithm. Windows mapping in the same chromosome region were used for graphical display.
Identification of centromeric regions by CenH3 ChIP
An antibody to CenH3 that showed specific binding to the cotton CenH3 protein was used in ChIP23. ChIP experiments were undertaken by following a published protocol65. High-quality Illumina reads were mapped to our cotton genome assembly with Bowtie 2 (http://bowtie-bio.sourceforge.net/bowtie2/index.shtml) as previously described18. The genome was then divided into 10-kb non-overlapping windows and uniquely mapping reads (allowing 1-bp mismatch between each read and the reference genome) were counted. The read density was calculated by dividing the total number of uniquely mapping reads by the total number of mapped nucleotides in each genomic window. To remove the impact of non-specific binding by rabbit serum, the read density was adjusted for background signal by using mock control data. CenH3 domains were identified with SICER v1.1 (ref. 66). We used 200-bp windows, required fold change/control of ≥5 and false-discovery rate (FDR) < 0.01, and allowed gaps of 400 bp when defining the CenH3 domains.
RNA-seq data generation
RNA was collected from the roots, stems, leaves and various reproductive organs of TM-1 and Hai7124 plants (details in Supplementary Table 47) and subjected to transcriptome sequencing, which yielded on average 25,530,023 reads for each replicate (three replicates for each sample) (Supplementary Table 47). Illumina reads were aligned to the reference TM-1 genome (v2.1) using TopHat 2.1.1 (ref. 67), and quantification of gene expression was performed with Cufflinks version 2.2.1 (http://cole-trapnell-lab.github.io/cufflinks/) by using the GTF annotation file.
PacBio sequencing and data analysis
For TM-1 and Hai7124, total RNA was extracted from the roots, stems and leaves of 2-week-old plants; from whole mature flowers; from ovules and fibers at 0 and 5 DPA; from ovules at 10 and 20 DPA; and from fibers at 10 and 20 DPA. PacBio-seq library preparation was performed as previously described68. In total, data were generated from 33 SMRT cells. Sequence data were processed with SMRTlink 4.0 software. PacBio transcriptome reads (867,006 for TM-1 and 879,582 for Hai7124) were used in gene annotation.
Gene prediction and annotation
Gene prediction and annotation were conducted as previously4. Proteins from seven plant genomes (Arabidopsis thaliana, Carica papaya, Hibiscus syriacus, T. cacao, G. raimondii, G. arboreum and Vitis vinifera) were downloaded from Phytozome (release 11)69 and the corresponding species database8,10. Protein sequences were aligned to the assembly using GenBlastA (version 1.0.4)70. GeneWise (version 2.4.1)71 was used to predict the exact gene structure of the corresponding genomic regions for each GenBlastA hit. Five ab initio gene prediction programs, Augustus (version 3.2.1)72, Genscan (version 1.0)73, GlimmerHMM (version 3.0.4)74, Geneid (version 1.4.4)75 and SNAP (version 2006-07-28)76, were used to predict coding regions in the repeat-masked genome. Finally, PacBio transcriptome reads were mapped to the assembly with BLAT (version 35)77 and RNA-seq data were mapped to the assembly with HISAT2 (version 2.0.1)78; StringTie (version 1.2.2)79 and TransDecoder (v1.1.1)80 were then used to assemble the transcripts and convert candidate coding regions into gene models. All gene models predicted with the above three approaches were combined by EvidenceModeler (EVM)80 into a non-redundant set of gene structures. The gene models were further filtered on the basis of their Cscore, protein coverage and coding sequence overlapping TEs, where Cscore is the BLASTP score ratio to the MBH (mutual best hit) BLASTP score and protein coverage is the highest percentage of the protein aligned to the best homolog. Only transcripts with Cscore ≥0.5 and protein coverage ≥0.5 were retained and the coding sequence overlapping repeats should be less than 20%; for gene models where more than 20% of the coding sequence overlapped repeats, the Cscore was required to be at least 0.9 and the protein coverage at least 70% for the model to be selected. Finally, gene models where more than 30% of the encoded protein was annotated as Pfam81 or InterPro82 TE domains were filtered out. Functional annotation of PCGs was achieved with BLASTP (version 2.2.26)83 (E value of 1 × 10–5) against two integrated protein sequence databases: SwissProt and TrEMBL84. Protein domains were annotated with InterProScan (v5.19)85. The GO terms for each gene were retrieved with Blast2GO86 based on the nr protein database. The pathways in which the genes might be involved were assigned by performing BLAST against the KEGG databases (release 59.3)87 (E value of 1 × 10–5).
Noncoding RNA annotation
tRNA-encoding genes were predicted by tRNAscan-SE (version 1.3.1)88. rRNA fragments were predicted by aligning to Arabidopsis and rice template rRNA sequences by using BLASTN (version 2.2.26) with an E-value cutoff of 1 × 10–10. MicroRNA and small nuclear RNA (snRNA) genes were found by searching against the Rfam database (release 12.0)89 with Infernal (version 1.1.1)90.
Repeat annotation
Repeats in the assembled genomes were probed with RepeatMasker (version open-4.0.6; http://www.repeatmasker.org/) run against the MIPS repeat database (mipsREdat_9.3p)91. RepeatMasker results were filtered to retain high-confidence hits (length ≥ 50 bp, score ≥ 255). To estimate the insertion times for LTRs, we applied LTR_FINDER (v1.06)92 to identify LTRs in the cotton genome with parameters ‘-D 15000 -d 1000 -L 7000 -l 100 -p 20 -C -M 0.9’ and then integrated the results and removed false positives from the initial predictions by the LTR_retriever pipeline93; insertion time was estimated as T = K/2r (where K is the divergence rate and r is the neutral mutation rate; r = 2.6 × 10−9) in the LTR_retriever package93.
Identification of homeologous and orthologous gene sets
A total of 9,282 one-to-one orthologous gene sets were identified for the G. arboreum, G. raimondii and T. cacao genomes and the two subgenomes of G. hirsutum and G. barbadense by using the OrthoMCL (version 1.4) clustering program94. 21,419 one-to-one orthologous gene sets were identified for G. raimondii, G. arboreum, and the two subgenomes of G. hirsutum and G. barbadense by using BLASTP (version 2.2.26)83 based on the bidirectional best hit (BBH) method with sequence coverage >30% and identity >30%, followed by selection of the best match.
Assessing the reference genome by BUSCO gene set
Quality control on the integrity of the assembly was performed by using the independent BUSCO v2 benchmark (http://busco.ezlab.org/)27, which is used to specifically assess the integrity of genic regions.
Phylogenetic tree construction and evolutionary rate estimation
A phylogenetic tree was constructed for the seven genomes and subgenomes (T. cacao, G. raimondii, G. arboreum, and the two subgenomes each from G. hirsutum and G. barbadense) by using coding sequence alignment of orthologs from these genomes. Evolutionary rates (Ka and Ks) on each branch were estimated by using the Codeml program in the PAML package (version 4.9d)95. We used the free-ratio ‘branch’ model, which allows distinct evolutionary rates for each branch. The significance of rate variations between linages was examined through Wilcoxon rank-sum tests.
Estimation of divergence time
On the basis of the 21,419 cotton orthologous gene sets for G. raimondii, G. arboreum, and the two subgenomes each of G. hirsutum and G. barbadense, the synonymous divergence levels (Ks) for all four cotton species were calculated. The formula t = Ks/2r was used to estimate the divergence time between species, where r is the neutral substitution rate (r = 2.6 × 10−9).
Phylogenetic tree of 13 D-genome diploids and 5 AD-genome allotetraploids
Seventeen accessions comprising 13 D-genome diploid species and 3 wild allotetraploid species, Gossypium tomentosum (AD3), Gossypium mustelinum (AD4) and Gossypium darwinii (AD5)96,97, were resequenced at high depth on the Illumina HiSeq 2500 platform, generating 85 Gb of sequencing data (Supplementary Table 16). All reads were trimmed for quality with Sickle by using a minimum Phred quality threshold of 20 (https://github.com/najoshi/sickle). SNPs were then called by aligning the high-quality reads from the 17 accessions, Hai7124 and the 3 wild allotetraploid species against the TM-1 (v2.1) reference genome. The phylogenetic tree was constructed with the neighbor-joining method98.
Gene family expansion and contraction
By using the gene families identified by the OrthoMCL (version 1.4) clustering program94, we compared the number of genes clustered in each family between TM-1 and Hai7124. If the number of genes in TM-1 was more than twofold higher than that in Hai7124 and there were at least five genes in the gene family or the gene family was specific to TM-1, these gene families were defined as families that were expanded in TM-1 and as families that were contracted in Hai7124, and vice versa.
Identification of SNPs, indels and structural variations between TM-1 and Hai7124
The Hai7124 (v1.1) genome was aligned with the TM-1 (v2.1) genome by MUMmer (version 3.23; http://mummer.sourceforge.net/) with parameters ‘-maxmatch 90 -l 40’, and one-to-one genomic alignment results were extracted with the ‘delta-filter -1’ parameter. SNPs and indels were identified by show-snp from the one-to-one alignment blocks (parameter ‘-ClrTH’). For identification of structural variations (PAVs, inversions and translocations), the alignment blocks from MUMmer with characteristics of structural variations were first extracted and the blocks with low similarity in the two flanking regions were filtered out.
To validate these variations, the following approaches were performed based on alignments of all available Illumina reads for TM-1 and Hai7124 to the TM-1 (v2.1) and Hai7124 (v1.1) genomes by BWA (version 0.7.15-r1140)62. In approach 1, at least four Illumina reads supporting the genotype were required to make us sufficiently confident of a candidate locus. In approach 2, Illumina reads were broken at the edge of the variation from the alignment results and variations with at least four supporting reads were defined as high-confidence variations. In approach 3, the single-end read/paired-end read ratio (S/P ratio) at the edge of the variation was computed and Fisher’s exact test was used to determine whether the variation was a supported candidate. SNPs were validated by approach 1. Indels (≤100 bp) were validated by approach 1 or 2. Structural variations (>100 bp; PAVs, inversions and translocations) were validated by approach 2 or 3. The putative functional effects of SNPs and indels were annotated with the ANNOVAR package99.
Phylogenetic relationship and genomic variations between G. hirsutum and G. barbadense
Nineteen representative cotton accessions were subjected to deep resequencing on the Illumina HiSeq X Ten platform, producing 3.0 Tb of raw data, corresponding to a median of 60-fold coverage for each sample (Supplementary Table 17). Paired-end resequencing reads from these accessions were mapped to the newly assembled TM-1 or Hai7124 genome by BWA62 (version 0.7.15-r1140) with the MEM algorithm. After alignment, all mapped reads were sorted according to their aligned chromosomal position and potential PCR duplicates were removed with SAMtools (version 1.3.1)100. SNP calling was performed with the GATK (version 3.7-0-gcfedb67) UnifiedGenotyper101 and SAMtools (version 1.3.1) mpileup100. To obtain high-quality SNPs, only variations detected with both software packages with a sequencing depth of at least 10 and Phred-scaled quality score of at least 30 were retained for further analysis. The evolutionary history of these accessions was inferred by using the neighbor-joining tree98.
By using the indel and structural variation superset between TM-1 and Hai7124, the genotype of the 19 accessions was identified with the above validation approach with Illumina reads. Indels (≤100 bp) were identified by approach 1 or 2. Structural variations (>100 bp; PAVs, inversions and translocations) were identified by approach 2 or 3.
CNVs were identified by CNVKit102 with gene annotations as targets. The segments identified with a log2-transformed copy number of at least 1.0 were considered to be copy number gain, and those with a log2-transformed copy number of −2.0 or less constituted to be copy number loss.
Sampling and gene expression profiles under abiotic stresses
Four-week-old seedlings were grouped in batches containing 15 seedlings each. Seedling batches were exposed to sodium chloride (0.4 M) and PEG (200 g/liter) as well as temperature regimes (Supplementary Table 47). Leaves were collected and used for transcriptome sequencing. Differentially expressed genes were identified by using the DESeq2 package (http://www.bioconductor.org/packages/release/bioc/html/DESeq2.html), and raw P values from multiple tests were corrected by using FDR. In identifying differentially expressed genes, we required at least a twofold change in expression and a P value of less than 0.05.
Quantitative RT–PCR analysis
Gene-specific primers were designed on the basis of the candidate genes with WebSNAPER (http://pga.mgh.harvard.edu/cgi-bin/snap3/websnaper3.cgi). qRT–PCR was performed with SYBR Green Mastermix (Vazyme) on an ABI7500 instrument according to the manufacturer’s instructions. Gene expression was normalized against expression of the GhEF1α gene. Relative transcript levels were computed by using the 2–ΔΔCt method on the basis of Ct values.
Statistical analysis
Comparison of codon substitution rate distributions between the two subgenomes of TM-1 and Hai7124 and their progenitors was carried out by using a two-sided Wilcoxon rank-sum test. Fisher’s exact test was employed to estimate whether a list of genes was enriched in a specific GO category when compared with background genes. P values were adjusted by FDR, and only GO terms with adjusted P < 0.05 were retained.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The TM-1 (v2.1) and Hai7124 (v1.1) assembly and annotation data are available at http://ibi.zju.edu.cn/cotton and http://www.cottongen.org/, respectively. The raw sequencing data used for de novo whole-genome assembly are available from the Sequence Read Archive under accession number PRJNA450479 and the China National GeneBank (CNGB) Nucleotide Sequence Archive (CNSA) under accession number CNP0000046. ChIP-seq data have been deposited in the Gene Expression Omnibus under accession number GSE119184. All raw data for the 19 deep-sequenced accessions including 10 G. hirsutum and 9 G. barbadense accessions are available under project number PRJNA450479, CNP0000046 and PRJNA375965. Illumina RNA-seq data are available at the Sequence Read Archive under accession number PRJNA490626. Hi-C data and Bionano data for TM-1 and Hai7124 are available at the Sequence Read Archive under accession number PRJNA505106. Further details on data accessibility are outlined in the supplementary materials and methods.
References
Doebley, J. F., Gaut, B. S. & Smith, B. D. The molecular genetics of crop domestication. Cell 127, 1309–1321 (2006).
Endrizzi, J., Turcotte, E. & Kohel, J. Genetics, cytogenetics and evolution of Gossypium. Adv. Genet. 23, 271–375 (1985).
Wendel, J. F. New World tetraploid cottons contain Old World cytoplasm. Proc. Natl Acad. Sci. USA 86, 4132–4136 (1989).
Zhang, T. et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 33, 531–537 (2015).
Liu, X. et al. Gossypium barbadense genome sequence provides insight into the evolution of extra-long staple fiber and specialized metabolites. Sci. Rep. 5, 14139 (2015).
Li, F. et al. Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat. Biotechnol. 33, 524–530 (2015).
Yuan, D. et al. The genome sequence of Sea-Island cotton (Gossypium barbadense) provides insights into the allopolyploidization and development of superior spinnable fibres. Sci. Rep. 5, 17662 (2015).
Du, X. et al. Resequencing of 243 diploid cotton accessions based on an updated A genome identifies the genetic basis of key agronomic traits. Nat. Genet. 50, 796–802 (2018).
Li, F. et al. Genome sequence of the cultivated cotton Gossypium arboreum. Nat. Genet. 46, 567–572 (2014).
Paterson, A. H. et al. Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492, 423–427 (2012).
Avni, R. et al. Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science 357, 93–97 (2017).
International Wheat Genome Sequencing Consortium. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361, eaar7191 (2018).
Luo, M. C. et al. Genome sequence of the progenitor of the wheat D genome Aegilops tauschii. Nature 551, 498–502 (2017).
Zhao, G. Y. et al. The Aegilops tauschii genome reveals multiple impacts of transposonsNat. Plants 3, 946–955 (2017).
Guo, L. et al. The opium poppy genome and morphinan production. Science 362, 343–347 (2018).
Springer, N. M. et al. The maize W22 genome provides a foundation for functional genomics and transposon biology. Nat. Genet. 50, 1282–1288 (2018).
Luo, S. et al. The cotton centromere contains a Ty3-gypsy-like LTR retroelement. PLoS One 7, e35261 (2012).
Su, H. et al. Dynamic location changes of Bub1-phosphorylated-H2AThr133 with CENH3 nucleosome in maize centromeric regions. New Phytol. 214, 682–694 (2017).
Jiang, J. & Birchler, J. A. Plant Centromere Biology (Wiley-Blackwell, 2013).
Schneider, K. L., Xie, Z., Wolfgruber, T. K. & Presting, G. G. Inbreeding drives maize centromere evolution. Proc. Natl Acad. Sci. USA 113, 987–996 (2016).
Wang, K., Wu, Y., Zhang, W., Dawe, R. K. & Jiang, J. Maize centromeres expand and adopt a uniform size in the genetic background of oat. Genome Res. 24, 107–116 (2014).
Gong, Z. et al. Repeatless and repeat-based centromeres in potato: implications for centromere evolution. Plant Cell 24, 3559–3574 (2012).
Han, J. et al. Rapid proliferation and nucleolar organizer targeting centromeric retrotransposons in cotton. Plant J. 88, 992–1005 (2016).
Zhu, Z. et al. The NnCenH3 protein and centromeric DNA sequence profiles of Nelumbo nucifera Gaertn (sacred lotus) reveal the DNA structures and dynamics of centromeres in basal eudicots. Plant J. 87, 568–582 (2016).
International Brachypodium Initiative. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463, 763–768 (2010).
Li, Y. et al. Centromeric DNA characterization in the model grass Brachypodium distachyon provides insights on the evolution of the genus. Plant J. 93, 1088–1101 (2018).
Simao, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V. & Zdobnov, E. M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212 (2015).
Wendel, J. F. & Cronn, R. C. Polyploidy and the evolutionary history of cotton. Adv. Agron. 78, 139–186 (2003).
Menzel, M. Y. & Brown, M. S. The significance of multivalent formation in three-species Gossypium hybrids. Genetics 39, 546–557 (1954).
Gerstel, D. U. Chromosomal translocations in interspecific hybrids of the genus Gossypium. Evolution 7, 11 (1953).
Brubaker, C. L., Paterson, A. H. & Wendel, J. F. Comparative genetic mapping of allotetraploid cotton and its diploid progenitors. Genome 42, 184–203 (1999).
Argout, X. et al. The genome of Theobroma cacao. Nat. Genet. 43, 101–108 (2011).
Fang, L. et al. Genomic insights into divergence and dual domestication of cultivated allotetraploid cottons. Genome Biol. 18, 33 (2017).
Westengen, O. T., Huaman, Z. & Heun, M. Genetic diversity and geographic pattern in early South American cotton domestication. Theor. Appl. Genet. 110, 392–402 (2005).
Han, L. B. et al. The dual functions of WLIM1a in cell elongation and secondary wall formation in developing cotton fibers. Plant Cell 25, 4421–4438 (2013).
Li, Y. et al. GbEXPATR, a species-specific expansin, enhances cotton fibre elongation through cell wall restructuring. Plant Biotechnol. J. 14, 951–963 (2016).
Andres, Z. et al. Control of vacuolar dynamics and regulation of stomatal aperture by tonoplast potassium uptake. Proc. Natl Acad. Sci. USA 111, E1806–E1814 (2014).
Ruan, Y. L., Llewellyn, D. J. & Furbank, R. T. The control of single-celled cotton fiber elongation by developmentally reversible gating of plasmodesmata and coordinated expression of sucrose and K+ transporters and expansin. Plant Cell 13, 47–60 (2001).
Barragan, V. et al. Ion exchangers NHX1 and NHX2 mediate active potassium uptake into vacuoles to regulate cell turgor and stomatal function in Arabidopsis. Plant Cell 24, 1127–1142 (2012).
Bassil, E. et al. The Arabidopsis Na+/H+ antiporters NHX1 and NHX2 control vacuolar pH and K+ homeostasis to regulate growth, flower development, and reproduction. Plant Cell 23, 3482–3497 (2011).
Hedrich, R., Sauer, N. & Neuhaus, H. E. Sugar transport across the plant vacuolar membrane: nature and regulation of carrier proteins. Curr. Opin. Plant Biol. 25, 63–70 (2015).
Meyer, S., De Angeli, A., Fernie, A. R. & Martinoia, E. Intra- and extra-cellular excretion of carboxylates. Trends Plant Sci. 15, 40–47 (2010).
Meyer, S. et al. Malate transport by the vacuolar AtALMT6 channel in guard cells is subject to multiple regulation. Plant J. 67, 247–257 (2011).
Nei, M. & Kumar, S. Molecular evolution and phylogenetics. Heredity 86, 385–385 (2000).
Wang, L. & Ruan, Y. L. Unraveling mechanisms of cell expansion linking solute transport, metabolism, plasmodesmtal gating and cell wall dynamics. Plant Signal. Behav. 5, 1561–1564 (2010).
Wang, L., Cook, A., Patrick, J. W., Chen, X. Y. & Ruan, Y. L. Silencing the vacuolar invertase gene GhVIN1 blocks cotton fiber initiation from the ovule epidermis, probably by suppressing a cohort of regulatory genes via sugar signaling. Plant J. 78, 686–696 (2014).
Wang, L. et al. Evidence that high activity of vacuolar invertase is required for cotton fiber and Arabidopsis root elongation through osmotic dependent and independent pathways, respectively. Plant Physiol. 154, 744–756 (2010).
Li, X. R., Wang, L. & Ruan, Y. L. Developmental and molecular physiological evidence for the role of phosphoenolpyruvate carboxylase in rapid cotton fibre elongation. J. Exp. Bot. 61, 287–295 (2010).
Zhang, Z. et al. Suppressing a putative sterol carrier gene reduces plasmodesmal permeability and activates sucrose transporter genes during cotton fiber elongation. Plant Cell 29, 2027–2046 (2017).
Naramoto, S. et al. ADP-ribosylation factor machinery mediates endocytosis in plant cells. Proc. Natl Acad. Sci. USA 107, 21890–21895 (2010).
Xu, J. & Scheres, B. Dissection of Arabidopsis ADP-RIBOSYLATION FACTOR 1 function in epidermal cell polarity. Plant Cell 17, 525–536 (2005).
Mittler, R., Finka, A. & Goloubinoff, P. How do plants feel the heat? Trends Biochem. Sci. 37, 118–125 (2012).
Kendrick, M. D. & Chang, C. Ethylene signaling: new levels of complexity and regulation. Curr. Opin. Plant Biol. 11, 479–485 (2008).
Raghavendra, A. S., Gonugunta, V. K., Christmann, A. & Grill, E. ABA perception and signalling. Trends Plant Sci. 15, 395–401 (2010).
Kohel, R., Richmond, T. & Lewis, C. Texas Marker-1. Description of a genetic standard for Gossypium hirsutum L. Crop Sci. 10, 670–671 (1970).
Paterson, A. H., Brubaker, C. L. & Wendel, J. F. A rapid method for extraction of cotton (Gossypium spp.) genomic DNA suitable for RFLP or PCR analysis. Plant Mol. Biol. Rep. 11, 122–127 (1993).
Zhang, M. et al. Preparation of megabase-sized DNA from a variety of organisms using the nuclei method for advanced genomics research. Nat. Protoc. 7, 467–478 (2012).
Wang, S. et al. Sequence-based ultra-dense genetic and physical maps reveal structural variations of allopolyploid cotton genomes. Genome Biol. 16, 108 (2015).
Van Ooijen, J. W. & Voorrips, R. JoinMap: version 3.0 (Plant Research International, 2001).
Wu, Y., Bhat, P. R., Close, T. J. & Lonardi, S. Efficient and accurate construction of genetic linkage maps from the minimum spanning tree of a graph. PLoS Genet. 4, e1000212 (2008).
Xie, T. et al. De novo plant genome assembly based on chromatin interactions: a case study of Arabidopsis thaliana. Mol. Plant 8, 489–492 (2015).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Akdemir, K. C. & Chin, L. HiCPlotter integrates genomic data with interaction matrices. Genome Biol. 16, 198 (2015).
Guo, W. et al. A preliminary analysis of genome structure and composition in Gossypium hirsutum. BMC Genomics 9, 314 (2008).
Nagaki, K. et al. Sequencing of a rice centromere uncovers active genes. Nat. Genet. 36, 138–145 (2004).
Zang, C. et al. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics 25, 1952–1958 (2009).
Kim, D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013).
Wang, B. et al. Unveiling the complexity of the maize transcriptome by single-molecule long-read sequencing. Nat. Commun. 7, 11708 (2016).
Goodstein, D. M. et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186 (2012).
She, R., Chu, J. S., Wang, K., Pei, J. & Chen, N. GenBlastA: enabling BLAST to identify homologous gene sequences. Genome Res. 19, 143–149 (2009).
Birney, E., Clamp, M. & Durbin, R. GeneWise and Genomewise. Genome Res. 14, 988–995 (2004).
Stanke, M., Schoffmann, O., Morgenstern, B. & Waack, S. Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinformatics 7, 62 (2006).
Burge, C. & Karlin, S. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268, 78–94 (1997).
Majoros, W. H., Pertea, M. & Salzberg, S. L. TigrScan and GlimmerHMM: two open source ab initio eukaryotic gene-finders. Bioinformatics 20, 2878–2879 (2004).
Guigo, R. Assembling genes from predicted exons in linear time with dynamic programming. J. Comput. Biol. 5, 681–702 (1998).
Korf, I. Gene finding in novel genomes. BMC Bioinformatics 5, 59 (2004).
Kent, W. J. Blat: the BLAST-like alignment tool. Genome Res. 12, 656–664 (2002).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015).
Pertea, M. et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295 (2015).
Haas, B. J. et al. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol. 9, R7 (2008).
Punta, M. et al. The Pfam protein families database. Nucleic Acids Res. 40, D290–D301 (2012).
Hunter, S. et al. InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 40, D306–D312 (2012).
Altschul, S. F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997).
Bairoch, A. & Apweiler, R. The SWISS-PROT protein sequence data bank and its supplement TrEMBL in 1999. Nucleic Acids Res. 27, 49–54 (1999).
Jones, P. et al. InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240 (2014).
Conesa, A. et al. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676 (2005).
Kanehisa, M. et al. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 42, D199–D205 (2014).
Lowe, T. M. & Eddy, S. R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964 (1997).
Griffiths-Jones, S. et al. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 33, D121–D124 (2005).
Nawrocki, E. P., Kolbe, D. L. & Eddy, S. R. Infernal 1.0: inference of RNA alignments. Bioinformatics 25, 1335–1337 (2009).
Nussbaumer, T. et al. MIPS PlantsDB: a database framework for comparative plant genome research. Nucleic Acids Res. 41, D1144–D1151 (2013).
Xu, Z. & Wang, H. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 35, W265–W268 (2007).
Ou, S. & Jiang, N. LTR_retriever: a highly accurate and sensitive program for identification of long terminal repeat retrotransposons. Plant Physiol. 176, 01310 (2017).
Li, L., Stoeckert, C. J. Jr. & Roos, D. S. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13, 2178–2189 (2003).
Yang, Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 (2007).
Fryxell, P. A. The Natural History of the Cotton Tribe (Texas A&M University Press, 1979).
Grover, C. E., Kim, H., Wing, R. A., Paterson, A. H. & Wendel, J. F. Incongruent patterns of local and global genome size evolution in cotton. Genome Res. 14, 1474–1482 (2004).
Saitou, N. & Nei, M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 (1987).
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Talevich, E. et al. CNVkit: genome-wide copy number detection and visualization from targeted DNA sequencing. PLoS Comput. Biol. 12, e1004873 (2016).
Acknowledgements
Research reported in this publication was supported by the National Key R&D Program for Crop Breeding (2016YFD0100505 to L.F., 2016YFD0101001 to J.C.), the National Science Foundation of China (31661143016 to T. Zhang), the Distinguished Discipline Support Program of Zhejiang University and the Esquel Group.
Author information
Authors and Affiliations
Contributions
T. Zhang and Y.H. conceptualized the project. Y.H., J.C., L.F., Z.Z., W.M. and T. Zhang planned and coordinated the project. Z.H., H.W., C.X., W.S. and D.Y. collected and grew the plant materials and prepared DNA and RNA for Illumina sequencing and PacBio SMRT sequencing. J.D., Z.H., Z.S., Y. Zang, X.Z. and B.Z. developed the F2 populations and prepared DNA for sequencing. K.B., A.B. and H.B. performed genome assembly. Y.N., J.L., L.Z. and F.H. conducted genome annotation and synteny analyses. J.C. constructed the genetic map and performed the structural variation study. Z.Z. and W.M. analyzed the genes related to fiber development and abiotic stresses. J.H. and K.W. identified centromeric regions by ChIP-seq. Q.W., L.J., T. Zhao, G.M., X.G., F.D., Y.B., X.L. and Y. Zhang were involved in data analysis. L.F., Y.N. and J.C. carried out data submission. Y.H., L.F., Z.Z., W.M., T. Zhang, M.R., Y.R., X.C., D.F. and S.Z. wrote the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figures 1–23 and Supplementary Tables 1–32, 36, 39–41 and 44–46
Supplementary Tables
Supplementary Tables 33–35, 37, 38, 42, 43 and 47
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hu, Y., Chen, J., Fang, L. et al. Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat Genet 51, 739–748 (2019). https://doi.org/10.1038/s41588-019-0371-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-019-0371-5
This article is cited by
-
Characterization and gene expression analysis reveal universal stress proteins respond to abiotic stress in Gossypium hirsutum
BMC Genomics (2024)
-
Genome-wide identification and expression profiling of photosystem II (PsbX) gene family in upland cotton (Gossypium hirsutum L)
Journal of Cotton Research (2024)
-
Transcriptomic datasets of Verticillium wilt resistant and non-resistant Gossypium barbadense varieties during pathogen inoculation
Scientific Data (2024)
-
Genetic improvement of Egyptian cotton (Gossypium barbadense L.) for high yield and fiber quality properties under semi arid conditions
Scientific Reports (2024)
-
Genome-wide identification of the key kinesin genes during fiber and boll development in upland cotton (Gossypium hirsutum L.)
Molecular Genetics and Genomics (2024)