Abstract
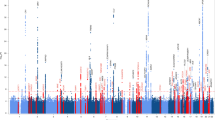
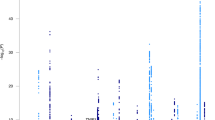
Risk for late-onset Alzheimer’s disease (LOAD), the most prevalent dementia, is partially driven by genetics. To identify LOAD risk loci, we performed a large genome-wide association meta-analysis of clinically diagnosed LOAD (94,437 individuals). We confirm 20 previous LOAD risk loci and identify five new genome-wide loci (IQCK, ACE, ADAM10, ADAMTS1, and WWOX), two of which (ADAM10, ACE) were identified in a recent genome-wide association (GWAS)-by-familial-proxy of Alzheimer’s or dementia. Fine-mapping of the human leukocyte antigen (HLA) region confirms the neurological and immune-mediated disease haplotype HLA-DR15 as a risk factor for LOAD. Pathway analysis implicates immunity, lipid metabolism, tau binding proteins, and amyloid precursor protein (APP) metabolism, showing that genetic variants affecting APP and Aβ processing are associated not only with early-onset autosomal dominant Alzheimer’s disease but also with LOAD. Analyses of risk genes and pathways show enrichment for rare variants (P = 1.32 × 10−7), indicating that additional rare variants remain to be identified. We also identify important genetic correlations between LOAD and traits such as family history of dementia and education.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
Genome-wide summary statistics for the Stage 1 discovery have been deposited in The National Institute on Aging Genetics of Alzheimer’s Disease Data Storage Site (NIAGADS)—a NIA/NIH-sanctioned qualified-access data repository, under accession NG00075. Stage 1 data (individual level) for the GERAD cohort can be accessed by applying directly to Cardiff University. Stage 1 ADGC data are deposited in NIAGADS. Stage 1 CHARGE data are accessible by applying to dbGaP for all US cohorts and to Erasmus University for Rotterdam data. AGES primary data are not available owing to Icelandic laws. Stage 2 and Stage 3 primary data are available upon request.
Change history
15 August 2019
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
References
Lambert, J. C. et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat. Genet. 45, 1452–1458 (2013).
Adams, P. M. et al. Assessment of the genetic variance of late-onset Alzheimer’s disease. Neurobiol. Aging. 41, 1–8 (2016).
Gatz, M. et al. Role of genes and environments for explaining Alzheimer disease. Arch. Gen. Psychiatry 63, 168–174 (2006).
Naj, A. C. et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat. Genet. 43, 436–441 (2011).
Seshadri, S. et al. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA 303, 1832–1840 (2010).
Hollingworth, P. et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat. Genet. 43, 429–435 (2011).
Jonsson, T. et al. Variant of TREM2 associated with the risk of Alzheimer’s disease. N. Engl. J. Med. 368, 107–116 (2013).
Guerreiro, R. et al. TREM2 variants in Alzheimer’s disease. N. Engl. J. Med. 368, 117–127 (2013).
Jun, G. et al. Meta-analysis confirms CR1, CLU, and PICALM as alzheimer disease risk loci and reveals interactions with APOE genotypes. Arch. Neurol. 67, 1473–1484 (2010).
Harold, D. et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat. Genet. 41, 1088–1093 (2009).
Lambert, J. C. et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat. Genet. 41, 1094–1099 (2009).
Zheng, J. et al. LD Hub: A centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics 33, 051094 (2017).
Bulik-Sullivan, B. K. et al. LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47, 291–295 (2015).
Sims, R. C. et al. Novel rare coding variants in PLCG2, ABI3 and TREM2 implicate microglial-mediated innate immunity in Alzheimer’s disease. Nat. Genet. 49, 1373–1387 (2017).
Liu, J. Z. et al. Case-control association mapping by proxy using family history of disease. Nat. Genet . 49, 325–331 (2017).
Desikan, R. S. et al. Polygenic overlap between c-reactive protein, plasma lipids, and Alzheimer’s disease. Circulation 131, 2061-2069 (2015).
Jun, G. R. et al. Transethnic genome-wide scan identifies novel Alzheimer’s disease loci. Alzheimers Dement. 13, 727–738 (2017).
Vassar, R. ADAM10 prodomain mutations cause late-onset Alzheimer’s disease: not just the latest FAD. Neuron 80, 250–253 (2013).
Kim, M. et al. Potential late-onset Alzheimer’s disease-associated mutations in the ADAM10 gene attenuate alpha-secretase activity. Hum. Mol. Genet. 18, 3987–3996 (2009).
Kehoe, P. G. et al. Variation in DCP1, encoding ACE, is associated with susceptibility to Alzheimer disease. Nat. Genet. 21, 71–72 (1999).
Meng, Y. et al. Association of polymorphisms in the Angiotensin-converting enzyme gene with Alzheimer disease in an Israeli Arab community. Am. J. Hum. Genet. 78, 871–877 (2006).
Lehmann, D. J. et al. Large meta-analysis establishes the ACE insertion-deletion polymorphism as a marker of Alzheimer’s disease. Am. J. Epidemiol. 162, 305–317 (2005).
Wang, X.-B. et al. Angiotensin-converting enzyme insertion/deletion polymorphism is not a major determining factor in the development of sporadic Alzheimer disease: evidence from an updated meta-analysis. PLoS ONE 9, e111406 (2014).
Cai, G. et al. Evidence against a role for rare ADAM10 mutations in sporadic Alzheimer disease. Neurobiol. Aging. 33, 416–417.e3 (2012).
Belbin, O. et al. A multi-center study of ACE and the risk of late-onset Alzheimer’s disease. J. Alzheimers. Dis. 24, 587–597 (2011).
Marioni, R. E. et al. GWAS on family history of Alzheimeras disease. Transl. Psychiatry 8, 99 (2018).
Yang, J. et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet. 44, 369–375 (2012).
Chang, J.-Y. & Chang, N.-S. WWOX dysfunction induces sequential aggregation of TRAPPC6AΔ, TIAF1, tau and amyloid β, and causes apoptosis. Cell Death Discov. 1, 15003 (2015).
Sze, C. I. et al. Down-regulation of WW domain-containing oxidoreductase induces tau phosphorylation in vitro: a potential role in Alzheimer’s disease. J. Biol. Chem. 279, 30498–30506 (2004).
Zhang, B. et al. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell 153, 707–720 (2013).
Bai, Z. et al. AlzBase: an integrative database for gene dysregulation in Alzheimer’s disease. Mol. Neurobiol. 53, 310–319 (2016).
Zhang, Y. et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 34, 11929–11947 (2014).
Zhang, Y. et al. Purification and characterization of progenitor and mature human astrocytes reveals transcriptional and functional differences with mouse. Neuron 89, 37–53 (2016).
Olah, M. et al. A transcriptomic atlas of aged human microglia. Nat. Commun. 9, 539 (2018).
Corder, E. H. et al. Protective effect of apolipoprotein E type 2 allele for late onset Alzheimer disease. Nat. Genet. 7, 180–184 (1994).
Kim, J., Basak, J. M. & Holtzman, D. M. The role of apolipoprotein E in Alzheimer’s disease. Neuron 63, 287–303 (2009).
Steinberg, S. et al. Loss-of-function variants in ABCA7 confer risk of Alzheimer’s disease. Nat. Genet. 47, 445–447 (2015).
Vasquez, J. B., Fardo, D. W. & Estus, S. ABCA7 expression is associated with Alzheimer’s disease polymorphism and disease status. Neurosci. Lett. 556, 58–62 (2013).
De Roeck, A. et al. An intronic VNTR affects splicing of ABCA7 and increases risk of Alzheimer’s disease. Acta Neuropathol. 135, 827–837 (2018).
De Roeck, A. et al. Deleterious ABCA7 mutations and transcript rescue mechanisms in early onset Alzheimer’s disease. Acta Neuropathol. 134, 475–487 (2017).
Chapuis, J. et al. Increased expression of BIN1 mediates Alzheimer genetic risk by modulating tau pathology. Mol. Psychiatry 18, 1225–1234 (2013).
Rogaeva, E. et al. The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer disease. Nat. Genet. 39, 168–177 (2007).
Vardarajan, B. N. et al. Coding mutations in SORL 1 and Alzheimer disease. Ann. Neurol. 77, 215–227 (2015).
Suh, J. et al. ADAM10 missense mutations potentiate beta-amyloid accumulation by impairing prodomain chaperone function. Neuron 80, 385–401 (2013).
Huang, K. et al. A common haplotype lowers PU.1 expression in myeloid cells and delays onset of Alzheimer’s disease. Nat. Neurosci. 20, 1052–1061 (2017).
Brouwers, N. et al. Alzheimer risk associated with a copy number variation in the complement receptor 1 increasing C3b/C4b binding sites. Mol. Psychiatry 17, 223–233 (2012).
Flister, M. J. et al. Identifying multiple causative genes at a single GWAS locus. Genome Res. 23, 1996–2002 (2013).
Farh, K. K.-H. et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature 518, 337–343 (2014).
Bis, J. C. et al. Whole exome sequencing study identifies novel rare and common Alzheimer’s-associated variants involved in immune response and transcriptional regulation. Mol. Psychiatry https://doi.org/10.1038/s41380-018-0112-7 (2018).
Vardarajan, B. N. et al. Coding mutations in SORL1 and Alzheimer disease. Ann. Neurol. 77, 215–227 (2015).
Verheijen, J. et al. A comprehensive study of the genetic impact of rare variants in SORL1 in European early-onset Alzheimer’s disease. Acta Neuropathol. 132, 213–224 (2016).
Bellenguez, C. et al. Contribution to Alzheimer’s disease risk of rare variants in TREM2, SORL1, and ABCA7 in 1779 cases and 1273 controls. Neurobiol. Aging. 59, 220.e1–220.e9 (2017).
Kunkle, B. W. et al. Targeted sequencing of ABCA7 identifies splicing, stop-gain and intronic risk variants for Alzheimer disease. Neurosci. Lett. 649, 124–129 (2017).
May, P. et al. Rare ABCA7 variants in 2 German families with Alzheimer disease. Neurol. Genet. 4, e224 (2018).
Guennec, K. Le et al. ABCA7 rare variants and Alzheimer disease risk. Neurology 86, 1–4 (2016).
Boyle, A. P. et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 22, 1790–1797 (2012).
Ward, L. D. & Kellis, M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 40, D930–D934 (2012).
Zerbino, D. R., Wilder, S. P., Johnson, N., Juettemann, T. & Flicek, P. R. The Ensembl Regulatory Build. Genome. Biol. 16, 56 (2015).
Huang, D. et al. GWAS4D: multidimensional analysis of context-specific regulatory variant for human complex diseases and traits. Nucleic Acids Res. 46, W114–W120 (2018).
Gjoneska, E. et al. Conserved epigenomic signals in mice and humans reveal immune basis of Alzheimer’s disease. Nature 518, 365–369 (2015).
de Leeuw, C. A., Mooij, J. M., Heskes, T. & Posthuma, D. MAGMA: Generalized Gene-Set Analysis of GWAS Data. PLoS Comput. Biol. 11, 1–19 (2015).
Stefanis, L. alpha-Synuclein in Parkinson’s disease. Cold Spring Harb. Perspect. Med. 2, 1–23 (2012).
Takeda, A. et al. C-terminal alpha-synuclein immunoreactivity in structures other than Lewy bodies in neurodegenerative disorders. Acta Neuropathol. 99, 296–304 (2000).
Campion, D., Pottier, C., Nicolas, G., Le Guennec, K. & Rovelet-Lecrux, A. Alzheimer disease: modeling an Aβ-centered biological network. Mol. Psychiatry 7, 861–871 (2016).
Yeh, F. L., Wang, Y., Tom, I., Gonzalez, L. C. & Sheng, M. TREM2 binds to apolipoproteins, including APOE and CLU/APOJ, and thereby facilitates uptake of amyloid-beta by microglia. Neuron 91, 328–340 (2016).
Fritsche, L. G. et al. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat. Genet. 48, 134–143 (2015).
Haass, C., Kaether, C., Thinakaran, G. & Sisodia, S. Trafficking and proteolytic processing of APP. Cold Spring Harb. Perspect. Med. 2, a006270 (2012).
Kleinberger, G. et al. TREM2 mutations implicated in neurodegeneration impair cell surface transport and phagocytosis. Sci. Transl. Med. 6, 243ra86 (2014).
Postina, R. et al. A disintegrin-metalloproteinase prevents amyloid plaque formation and hippocampal defects in an Alzheimer disease mouse model. J. Clin. Invest. 113, 1456–1464 (2004).
Hinney, A. et al. Genetic variation at the CELF1 (CUGBP, elav-like family member 1 gene) locus is genome-wide associated with Alzheimer’s disease and obesity. Am. J. Med. Genet. B. 165B, 283–293 (2014).
Speliotes, E. K. et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat. Genet. 42, 937–948 (2010).
Kurabayashi, N., Nguyen, M. D. & Sanada, K. The G protein-coupled receptor GPRC5B contributes to neurogenesis in the developing mouse neocortex. Development 140, 4335–4346 (2013).
Cool, B. H. et al. A flanking gene problem leads to the discovery of a Gprc5b splice variant predominantly expressed in C57BL/6J mouse brain and in maturing neurons. PLoS ONE 5, e10351 (2010).
Kim, Y.-J., Sano, T., Nabetani, T., Asano, Y. & Hirabayashi, Y. GPRC5B activates obesity-associated inflammatory signaling in adipocytes. Sci. Signal. 5, ra85–ra85 (2012).
Bhat, K. et al. The 19S proteasome ATPase Sug1 plays a critical role in regulating MHC class II transcription. Mol. Immunol. 45, 2214–2224 (2008).
Inostroza-Nieves, Y., Venkatraman, P. & Zavala-Ruiz, Z. Role of Sug1, a 19S proteasome ATPase, in the transcription of MHC I and the atypical MHC II molecules, HLA-DM and HLA-DO. Immunol. Lett. 147, 67–74 (2012).
Kim, K., Duramad, O., Qin, X. F. & Su, B. MEKK3 is essential for lipopolysaccharide-induced interleukin-6 and granulocyte-macrophage colony-stimulating factor production in macrophages. Immunology 120, 242–250 (2007).
Yamazaki, K. et al. Two mechanistically and temporally distinct NF-κB activation pathways in IL-1 signaling. Sci. Signal. 2, 1–12 (2009).
Farrer, L. A. et al. Association between angiotensin-converting enzyme and Alzheimer disease. New Engl. J. Med. 57, 210–214 (2000).
Miners, J. S. et al. Angiotensin-converting enzyme levels and activity in Alzheimer’s disease: differences in brain and CSF ACE and association with ACE1 genotypes. Am. J. Transl. Res. 1, 163–177 (2009).
Jochemsen, H. M. et al. The association of angiotensin-converting enzyme with biomarkers for Alzheimer’s disease. Alzheimers Res. Ther. 6, 1–10 (2014).
Kauwe, J. S. K. et al. Genome-wide association study of CSFl Levels of 59 Alzheimer’s disease candidate proteins: significant associations with proteins involved in amyloid processing and inflammation. PLoS Genet. 10, e1004758 (2014).
Baranello, R. J. et al. Amyloid-beta protein clearance and degradation (ABCD) pathways and their role in Alzheimer’s disease. Curr. Alzheimers Res 12, 32–46 (2015).
Kehoe, P. G. The coming of age of the angiotensin hypothesis in Alzheimer’s disease: progress toward disease prevention and treatment? J. Alzheimers. Dis. 62, 1443–1466 (2018).
Kehoe, P. G. et al. The rationale and design of the reducing pathology in Alzheimer’s disease through Angiotensin TaRgeting (RADAR) Trial. J. Alzheimers. Dis. 61, 803–814 (2017).
Miguel, R. F., Pollak, A. & Lubec, G. Metalloproteinase ADAMTS-1 but not ADAMTS-5 is manifold overexpressed in neurodegenerative disorders as Down syndrome, Alzheimer’s and Pick’s disease. Brain. Res. Mol. Brain. Res. 133, 1–5 (2005).
Suttkus, A. et al. Aggrecan, link protein and tenascin-R are essential components of the perineuronal net to protect neurons against iron-induced oxidative stress. Cell Death Dis. 5, e1119 (2014).
Végh, M. J. et al. Reducing hippocampal extracellular matrix reverses early memory deficits in a mouse model of Alzheimer’s disease. Acta Neuropathol. Commun. 2, 76 (2014).
Morawski, M., Filippov, M., Tzinia, A., Tsilibary, E. & Vargova, L. ECM in brain aging and dementia. Prog. Brain. Res. 214, 207–227 (2014).
Wilcock, D. M. Neuroinflammation in the aging down syndrome brain; lessons from Alzheimer’s disease. Curr. Gerontol. Geriatr. Res. 2012, 170276 (2012).
Wang, K. et al. A genome-wide association study on obesity and obesity-related traits. PLoS ONE 6, 3–8 (2011).
Kang, K. et al. Interferon-γ represses M2 gene expression in human macrophages by disassembling enhancers bound by the transcription factor MAF. Immunity 47, 235–250.e4 (2017).
Cao, S., Liu, J., Song, L. & Ma, X. The protooncogene c-Maf Is an essential transcription factor for IL-10 gene expression in macrophages. J. Immunol. 174, 3484–3492 (2005).
Lee, J. C. et al. WW-domain-containing oxidoreductase is associated with low plasma HDL-C levels. Am. J. Hum. Genet. 83, 180–192 (2008).
Saez, M. E. et al. WWOX gene is associated with HDL cholesterol and triglyceride levels. BMC. Med. Genet. 11, 148 (2010).
Chang, H. T. et al. WW domain-containing oxidoreductase in neuronal injury and neurological diseases. Oncotarget 5, 11792–11799 (2014).
Lee, M. H. et al. Zfra restores memory deficits in Alzheimer’s disease triple-transgenic mice by blocking aggregation of TRAPPC6AΔ, SH3GLB2, tau, and amyloid β, and inflammatory NF-κB activation. Alzheimers Dement. Transl. Res. Clin. Interv 3, 189–204 (2017).
Dourlen, P. et al. Functional screening of Alzheimer risk loci identifies PTK2B as an in vivo modulator and early marker of Tau pathology. Mol. Psychiatry 22, 874–883 (2017).
Chapuis, J. et al. Genome-wide, high-content siRNA screening identifies the Alzheimer’s genetic risk factor FERMT2 as a major modulator of APP metabolism. Acta Neuropathol. 133, 955–966 (2017).
Shulman, J. M. et al. Functional screening in Drosophila identifies Alzheimer’s disease susceptibility genes and implicates tau-mediated mechanisms. Hum. Mol. Genet. 23, 870–877 (2014).
Zhao, Z. et al. Central role for PICALM in amyloid-β blood-brain barrier transcytosis and clearance. Nat. Neurosci. 18, 978–987 (2015).
Aguet, F. et al. Genetic effects on gene expression across human tissues. Nature 550, 204–213 (2017).
Miller, J. A. et al. Transcriptional landscape of the prenatal human brain. Nature 508, 199–206 (2014).
Knickmeyer, R. C. & Ross, M. E. Imaging and rare APOE alleles. Neurology 87, 558–559 (2016).
Douaud, G. et al. A common brain network links development, aging, and vulnerability to disease. Proc. Natl Acad. Sci. USA 111, 17648–17653 (2014).
Steele, N. Z. et al. Fine-mapping of the human leukocyte antigen locus as a risk factor for Alzheimer disease: a case-control study. PLoS Med. 14, 1–25 (2017).
Fekih Mrissa, N. et al. Association of HLA-DR-DQ polymorphisms with diabetes in Tunisian patients. Transfus. Apher. Sci. 49, 200–204 (2013).
Pugliese, A. et al. HLA-DRB1 15:01-DQA1 01:02-DQB1 06:02 haplotype protects autoantibody-positive relatives from type 1 diabetes throughout the stages of disease progression. Diabetes 65, 1109–1119 (2016).
Patsopoulos, Na et al. Fine-mapping the genetic association of the major histocompatibility complex in multiple sclerosis: HLA and non-HLA effects. PLoS Genet. 9, e1003926 (2013).
Schmidt, H., Williamson, D. & Ashley-Koch, A. HLA-DR15 haplotype and multiple sclerosis: a HuGE review. Am. J. Epidemiol. 165, 1097–1109 (2007).
Karnes, J. H. et al. Phenome-wide scanning identifies multiple diseases and disease severity phenotypes associated with HLA variants. Sci. Transl. Med. 9, 1–14 (2017).
Wissemann, W. T. et al. Association of Parkinson disease with structural and regulatory variants in the HLA region. Am. J. Hum. Genet. 93, 984–993 (2013).
Misra, M. K., Damotte, V. & Hollenbach, J. A. The immunogenetics of neurological disease. Immunology 153, 399–414 (2018).
Tan, Z. S. Thyroid function and the risk of Alzheimer disease: the Framingham study. Arch. Intern. Med. 168, 1514 (2008).
Dendrou, C. A., Petersen, J., Rossjohn, J. & Fugger, L. HLA variation and disease. Nat. Rev. Immunol. 18, 325–339 (2018).
Bulik-Sullivan, B. et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 47, 1236–1241 (2015).
Stern, Y. Cognitive reserve in ageing and Alzheimer’s disease. Lancet. Neurol. 11, 1006–1012 (2012).
Cadar, D. et al. Individual and area-based socioeconomic factors associated with dementia incidence in England: evidencefrom a 12-year follow-up in the English longitudinal study of ageing. JAMA Psychiatry 75, 723–732 (2018).
Marden, J. R., Tchetgen Tchetgen, E. J., Kawachi, I. & Glymour, M. M. Contribution of socioeconomic status at 3 life-course periods to late-life memory function and decline: early and late predictors of dementia risk. Am. J. Epidemiol. 186, 805–814 (2017).
Østergaard, S. D. S. D. et al. Associations between potentially modifiable risk factors and Alzheimer disease: a Mendelian randomization study. PLoS Med. 12, e1001841 (2015).
Zhu, Z. et al. Causal associations between risk factors and common diseases inferred from GWAS summary data. Nat. Commun. 9, 224 (2018).
Baumgart, M. et al. Summary of the evidence on modifiable risk factors for cognitive decline and dementia: a population-based perspective. Alzheimers Dement. 11, 1–9 (2015).
Larsson, S. C., Traylor, M., Burgess, S. & Markus, H. S. Genetically-predicted adult height and Alzheimer’s disease. J. Alzheimers. Dis. 60, 691–698 (2017).
Helzner, E. P. et al. Contribution of vascular risk factors to the progression in Alzheimer disease. Arch. Neurol. 66, 343–348 (2009).
Reitz, C. et al. Association of higher levels of high-density lipoprotein cholesterol in elderly individuals and lower risk of late-onset Alzheimer disease. Arch. Neurol. 67, 1491–1497 (2010).
Mukherjee, S. et al. Genetically predicted body mass index and Alzheimer’s disease-related phenotypes in three large samples: Mendelian randomization analyses. Alzheimers Dement. 11, (2015).
Arvanitakis, Z. et al. Late-life blood pressure association with cerebrovascular and Alzheimer disease pathology. Neurology 91, e517–e525 (2018).
Kuźma, E. et al. Which risk factors causally influence dementia? A systematic review of mendelian randomization studies. J. Alzheimers. Dis. 36, 215–221 (2018).
Murray, M. E. et al. Clinicopathologic and 11C-Pittsburgh compound B implications of Thal amyloid phase across the Alzheimer’s disease spectrum. Brain 138, 1370–1381 (2015).
Shi, Y. et al. ApoE4 markedly exacerbates tau-mediated neurodegeneration in a mouse model of tauopathy. Nature 549, 523–527 (2017).
Brier, M. R. M. R. et al. Tau and Aβ imaging, CSF measures, and cognition in Alzheimer’s disease. Sci. Transl. Med. 8, 338ra66 (2016).
Genomes Project, C.. et al. An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65 (2012).
Howie, B. N., Donnelly, P. & Marchini, J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 5, e1000529 (2009).
Delaneau, O., Marchini, J. & Zagury, J. F. A linear complexity phasing method for thousands of genomes. Nat. Methods 9, 179–181 (2012).
Li, Y., Willer, C. J., Ding, J., Scheet, P. & Abecasis, G. R. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes.. Genet. Epidemiol. 34, 816–834 (2010).
Howie, B., Fuchsberger, C., Stephens, M., Marchini, J. & Abecasis, G. R. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat. Genet. 44, 955–959 (2012).
Howie, B., Marchini, J. & Stephens, M. Genotype imputation with thousands of genomes. G3 1, 457–470 (2011).
Price, A. L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006).
Ma, C. et al. Recommended joint and meta-analysis strategies for case-control association testing of single low-count variants. Genet. Epidemiol. 37, 539–550 (2013).
Chen, M.-H. H. & Yang, Q. GWAF: an R package for genome-wide association analyses with family data. Bioinformatics 26, 580–581 (2010).
Willer, C. J., Li, Y. & Abecasis, G. R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191 (2010).
Aulchenko, Y. S., Ripke, S., Isaacs, A. & van Duijn, C. M. GenABEL: an R library for genome-wide association analysis. Bioinformatics 23, 1294–1296 (2007).
Sudlow, C. et al. UK Biobank: An open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 12, 1–10 (2015).
Fry, A. et al. Comparison of sociodemographic and health-related characteristics of UK Biobank participants with those of the general population. Am. J. Epidemiol. 186, 1026–1034 (2017).
Patterson, N., Price, A. L. & Reich, D. Population structure and eigenanalysis. PLoS Genet. 2, e190 (2006).
Machiela, M. J. & Chanock, S. J. LDlink: A web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics 31, 3555–3557 (2015).
Zhang, X. et al. Synthesis of 53 tissue and cell line expression QTL datasets reveals master eQTLs. BMC Genomics 15, 532 (2014).
Pruitt, K. D., Tatusova, T., Brown, G. R. & Maglott, D. R. NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res. 40, D130–D135 (2012).
Harrow, J. et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 22, 1760–1774 (2012).
Arnold, M., Raffler, J., Pfeufer, a, Suhre, K. & Kastenmuller, G. SNiPA: an interactive, genetic variant-centered annotation browser. Bioinformatics 31, 1334–1336 (2014).
McLaren, W. et al. The Ensembl variant effect predictor. Genome Biol. 17, 122 (2016).
Cargill, M. et al. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat. Genet. 22, 231–238 (1999).
Ng, P. C. & Henikoff, S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 31, 3812–3814 (2003).
Adzhubei, I. A. et al. A method and server for predicting damaging missense mutations. Nat. Methods 7, 248–249 (2010).
Kircher, M. et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 46, 310–315 (2014).
Gonzalez-Perez, A. & Lopez-Bigas, N. Improving the assessment of the outcome of nonsynonymous SNVs with a consensus deleteriousness score, Condel. Am. J. Hum. Genet. 88, 440–449 (2011).
Samocha, K. E. et al. Regional missense constraint improves variant deleteriousness prediction. Preprint at https://doi.org/10.1101/148353 (2017).
Ionita-Laza, I., McCallum, K., Xu, B. & Buxbaum, J. D. A spectral approach integrating functional genomic annotations for coding and noncoding variants. Nat. Genet. 48, 214–220 (2016).
Yeo, G. & Burge, C. B. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol. 11, 377–394 (2004).
Amlie-Wolf, A. et al. INFERNO—INFERring the molecular mechanisms of NOncoding genetic variants. Nucleic Acids Res. 46, 8740–8753 (2018).
Ward, L. D. & Kellis, M. HaploRegv4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 44, D877–D881 (2015).
Thériault, P., ElAli, A. & Rivest, S. The dynamics of monocytes and microglia in Alzheimer’s disease. Alzheimers Res. Ther. 7, 41 (2015).
Raj, T. et al. Polarization of the effects of autoimmune and neurodegenerative risk alleles in leukocytes. Science 344, 519–523 (2014).
Qi, T. et al. Identifying gene targets for brain-related traits using transcriptomic and methylomic data from blood. Nat. Commun. 9, 2282 (2018).
Schramm, K. et al. Mapping the genetic architecture of gene regulation in whole blood. PLoS ONE 9, e93844 (2014).
Giambartolomei, C. et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 10, e1004383 (2014).
Zhu, Z. et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet. 48, 481–489 (2016).
Westra, H.-J. et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat. Genet. 45, 1238–1243 (2013).
Ashburner, M. et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29 (2000).
Blake, J. A. et al. Gene ontology consortium: going forward. Nucleic Acids Res. 43, D1049–D1056 (2015).
Kanehisa, M., Sato, Y., Kawashima, M., Furumichi, M. & Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44, D457–D462 (2016).
Ogata, H. et al. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 27, 29–34 (1999).
Fabregat, A. et al. The reactome pathway knowledgebase. Nucleic Acids Res. 44, D481–D487 (2016).
Croft, D. et al. Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res. 39, D691–D697 (2011).
Eppig, J. T., Blake, Ja, Bult, C. J., Kadin, Ja & Richardson, J. E. The Mouse Genome Database (MGD): facilitating mouse as a model for human biology and disease. Nucleic Acids Res. 43, D726–D736 (2014).
O’Dushlaine, C. et al. Psychiatric genome-wide association study analyses implicate neuronal, immune and histone pathways. Nat. Neurosci. 18, 199–209 (2015).
Szklarczyk, D. et al. STRINGv10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43, D447–D452 (2015).
Pletscher-Frankild, S., Pallejà, A., Tsafou, K., Binder, J. X. & Jensen, L. J. DISEASES: text mining and data integration of disease-gene associations. Methods 74, 83–89 (2015).
Santos, A. et al. Comprehensive comparison of large-scale tissue expression datasets. PeerJ 3, e1054 (2015).
Lachmann, A. et al. Massive mining of publicly available RNA-seq data from human and mouse. Nat. Commun. 9, 1366 (2018).
Kuleshov, M. V. et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97 (2016).
Watanabe, K., Taskesen, E., Van Bochoven, A. & Posthuma, D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 8, 1826 (2017).
Zheng, X. et al. HIBAG—HLA genotype imputation with attribute bagging. Pharmacogenomics. J. 14, 192–200 (2014).
R v.3.4.3 (R Development Core Team, 2017).
haplo.stats v.1.7.9 (2018).
Acknowledgements
We thank all the participants of this study for their contributions. Additional acknowledgements and detailed acknowledgments of funding sources for the study are provided in the Supplementary Note.
Author information
Authors and Affiliations
Consortia
Contributions
ADGC. Study design or conception: A.C.N., A.A.-W., E.R.M., K.H.-N., A.B.K., B.N.V., G.W.B., O.V., M.Butkiewicz, W.B., Y.Song, G.D.S., M.A.P.-V. Sample contribution: S.Mukherjee, P.K.C., R.B., P.M.A., M.S.A., D. Beekly, D. Blacker, R.S. Doody, T.J.F., M.P.F., B.Ghetti, R.M.H., M.I.K., M.J.K., C.K., W.K., E.B.L., R.B.L., T.J.M., R.C.P., E.M.R., J.S.R., D.R.R., M. Sano, P.S.G.-H., D.W.T., C.K.W., R.L.A., L.G.A., S.E.A., S.A., C.S.A., C.T.B., L.L.B., S. Barral, T.G.B., J.T.B., E.H.B., T.D.B., B.F.B., J.D.B., A.Boxer, J.R.B., J.M.B., J.D.Buxbaum, N.J.C., C. Cao, C.S.C., C.M.C., R.M.C., H.C.C., D.H.C., E.A.C., C.DeCarli, M.Dick, R.D., N.R.G.-R., D.A.E., K.M.F., K.B.F., D.W.F., M.R.F., S.F., T.M.F., D.R.G., M.Gearing, D.H.G., J.R.G., R.C.G., J.H.G., R.L.H., L.E.H., L.S.H., M.J.H., C.M.H., B.T.H., G.P.J., E.A., L.W.J., G.R.J., A. Karydas, J.A.K., R.K., N.W.K., J.H.K., F.M.L., J.J.L., J.B.L., A.I.L., A.P.L., K.L.L., C.G.L., D.C.M., F.M., D.C.Mash, E.M., W.C.M., S.M.M., A.N.M., A.C.M., M.M., B.L.M., C.A.M., J.W.M., J.C.M., A.J.M., S.O., J.M.O., J.E.P., H.L.P., E.P., A.P., W.W.P., H.P., J.F.Q., A.Raj, M.R., B.R., C.R., J.M.R., E.D.R., E.R., H.J.R., R.N.R., M.A.S., A.J.S., M.L.C., J. Vance, J.A.S., L.S.S., W.W.S., A.G.S., J.A.Sonnen, S. Spina, R.A.S., R.H.S., R.E.T., J.Q.T., J.C.T., V.M.V.D., L.J.V.E., H.V.V., J.P.V., S.W., K.A.W.-B., K.C.W., J.Williamson, T.S.W., R.L.W., C.B.W., C.-E.Y., L.Y., D.B., P.L.D.J., C.Cruchaga, A.M.G., N.E.-T., S.G.Y., D.W.D., H.H., L.A.F., J.Haines, R.Mayeux, L.-S.W., G.D.S., M.A.P.-V. Data generation: B.W.K., K.H.-N., A.B.K., O.V., L.Q., Y.Z., W.P., S.Slifer, J.Malamon, B.A.D., P.W., L.B.C., M.A., M.Tang, J.R.G., L.-S.W. Analysis: B.W.K., A.C.N., A.A.-W., E.R.M., K.H.-N., A.B.K., M.Tang, M.M.C., B.N.V., G.W.B., O.V., M.Butkiewicz, W.B., Y.S., G.D.S., M.A.P.-V. Manuscript preparation: B.W.K., G.D.S., M.A.P.-V. Study supervision/management: B.W.K., L.A.F., J.Haines, R.Mayeux, L.-S.W., G.D.S., M.A.P.-V. EADI. Study design or conception: P.A., J.-C.L. Sample contribution: K.S., M.Hiltunen, J.E., M.D.Z., I.M., F.S.-G., M.C.D.N., D.Wallon, S.E., R.V., P.D.D., A.Squassina, E.R.-R., C.M.-F., Y.A.B., H.T., V.Giedraitis, L.Kilander, R.Brundin, L.C., S.Helisalmi, A.M.K., A.Haapasalo, V.S., V.Frisardi, V.Deramecourt, N.F., O.H., C.Dufouil, A.Brice, K.R., B.D., H.Soininen, L.Fratiglioni, L.K., F.Panza, D.H., P.C., F.S., P.B., L.Lannfelt, F.P., M.Ingelsson, C.G., P.S.-J., A.L., J.Clarimon, C.Berr, S.D., J.-F.D., A.Pilotto, M.J.B., P.Bosco, E.C., G.N., D.C., C.V.B., P.A., J.-C.L. Data generation: R.O., J.-G.G., M.-L.M., D.Bacq, F.G., B.F., S.Meslage Analysis: B.G.-B., V.D., C.Bellenguez Manuscript preparation: B.G.-B., P.A., J.-C.L. Study supervision/management: J.-F.Deleuze, A.Boland, P.A., J.-C.L. GERAD/PERADES. Study design or conception: R.Sims, M.C.O., M.J.O., A.R., P.A.H., J.W. Sample contribution: R.Raybould, T.Morgan, P.Hoffmann, D.Harold, O.P., N.D., N.C.F., J.T.H., Y.P., M.Daniilidou, J.U., D.Galimberti, E.Scarpini, J.Kornhuber, S.Sordon., M.Mayhaus, W.G., A.M.H., S.Lovestone, R.Sussams, C.Holmes, W.M., A.Kawalia, S.Moebus, J.Turton, J.Lord, I.K., A.L., B.L., M.Gill, M.D.-F., I.A., A.Ciaramella, C.Cupidi, R.G.M., R.Cecchetti, M.T., D.Craig, D.A., A.G., M.K., O.G., H.Hampel, D.C.R., L.F., B.M., J.A.J., P.Passmore, J.M.S., J.D.W., M.K.L., P.Proitsi, J.Powell, J.S.K.K., M.Mancuso, U.B., A.M., G.Livingston, N.J.B., J.Hardy, J.B., R.Guerreiro, E.F., C.Masullo, G.B., L.M., A.H., M.Scherer, M.Riemenschneider, R.Heun, H.K., M.Leber, I.H., I.G., M.Hull, J.M., K.Mayo, T.F., D.Drichel, T.D.C., P.Hollingworth, R.Marshall, A.Meggy, G.M., G.L., D.G., G.R., F.J., B.V., E.V., K.-H.J., M.Dichgans, D.Mann, S.P.-B., N.K., H.W., K.M., K.Brown, C.Medway, M.M.N., N.M.H., A.Daniele, A.Bayer, J.G., H.V.D.B., C.Brayne, S.R.-H., A.A.-C., C.E.S., J.Wiltfang, V.A., A.B.S., J.C., S.M., M.Rossor, N.S.R., B.N., S.Sorbi, E.S., G.S., C.Caltagirone, M.D.O., R.C., A.D.S., D.W., G.W., A.C.B., M.G., Y.B.-S., P.M., P.P., V.B., N.W., P.D., R.G., P.G.K., S.L., C.C., J.T., R.Munger, A.R., J.W. Data generation: R.Sims, R.Raybould, T.Morgan, P.Hoffmann, D.Harold, A.Gerrish, N.D., P.Hollingworth, R.Marshall, A.Meggy, A.R., J.W. Analysis: R.Sims, M.V., A.F., N.Badarinarayan, D.Harold, G.M., G.L., D.G., V.E.-P., A.R., J.W., P.A.H. Manuscript preparation: R.Sims, T.D.C., P.A.H., J.W. Study supervision/management: R.Sims, L.J., V.E.-P., A.R., P.A.H., J.W. CHARGE. Study design or conception: A.L.D., C.M.V.D., S.S. Sample contribution: J.C.B., A.Ruiz, I.D.R., L.M.R., I.Q., A.C., A.L.F., G.E., J.J.H., A.O., M.E.G., H.L., H.Comic, G.Roshchupkin, S.Li, I.Hernández, Q.Y., A.S.B., L.T., T.H.M., WT.L., F.R., E.Boerwinkle, J.I.R., A.G.U., S.M.-G., O.L.L., M.B., M.F., N.A., L.J.L., M.A.I., H.S., R.S., V.G., B.M.P. Data generation: J.C.B., J.Jakobsdottir, A.Ruiz, A.V.S., X.J., S.-H.C., H.H.A., J.A.B., T.A., E.H., C.Sarnowski, D.V., L.A.C. Analysis: J.C.B., S.J.v.d.L., V.C., J.Jakobsdottir, Y.C., Y.Saba, S.Ahmad, A.Ruiz, A.V.S., C.C.W., C.M.V.D., S.S. Manuscript preparation: S.J.v.d.L., A.Ruiz, B.M.P., C.M.V.D., S.S. Study supervision/management: C.M.V.D., S.S.
Corresponding authors
Ethics declarations
Competing interests
D. Blacker is a consultant for Biogen, Inc. R.C.P. is a consultant for Roche, Inc.; Merck, Inc.; Genentech, Inc.; Biogen, Inc.; GE Healthcare; and Eisai, Inc. A.R.W. is a former employee and stockholder of Pfizer, Inc., and a current employee of the Perelman School of Medicine at the University of Pennsylvania Orphan Disease Center in partnership with the Loulou. A.M.G. is a member of the scientific advisory board for Denali Therapeutics. N.E.-T. is a consultant for Cytox. J.Hardy holds a collaborative grant with Cytox cofunded by the Department of Business (Biz). F.J. acts as a consultant for Novartis, Eli Lilly, Nutricia, MSD, Roche and Piramal. Neither J.M. nor his family owns stock or has equity interest (outside of mutual funds or other externally directed accounts) in any pharmaceutical or biotechnology company. J.M. is currently participating in clinical trials of antidementia drugs from Eli Lilly and Company, Biogen and Janssen. J.M. serves as a consultant for Lilly USA. He receives research support from Eli Lilly/Avid Radiopharmaceuticals and is funded by NIH grant nos. P50AG005681, P01AG003991, P01AG026276 and UF01AG032438. C.Cruchaga receives research support from Biogen, EISAI, Alector and Parabon. The funders of the study had no role in the collection, analysis or interpretation of data; in the writing of the report; or in the decision to submit the paper for publication. C.Cruchaga is a member of the advisory board of ADx Healthcare. M.R.F. receives grant/research support from AbbVie, Accera, ADCS Posiphen, Biogen, Eisai, Eli Lilly, Genentech, Novartis and Suven Life Sciences, Ltd. He is a consultant/advisory board/DSMB board member for Accera, Avanir, AZTherapies, Cognition Therapeutics, Cortexyme, Eli Lilly & Company, Longeveron, Medavante, Merck and Co. Inc., Otsuka Pharmaceutical, Proclara (formerly Neurophage Pharmaceuticals), Neurotrope Biosciences, Takeda, vTv Therapeutics and Zhejian Hisun Pharmaceuticals. He has a transgenic mouse model patent that is licensed to Elan. R.A.S. receives consulting fees as a member of the Alzheimer’s Disease Advisory Board, Biogen; and as member of the Executive Committee for AZD3293 Alzheimer’s Disease Studies, Eli Lilly. R.B.L. receives consulting fees from Merch, Inc. E.M.R. receives grant funding from several NIH grant and research contracts with Genentech/Roche, Novartis/Amgen and Avid/Lilly. He is a compensated scientific advisor to Alkahest, Alzheon, Aural Analytics, Denali, Takeda and Zinfandel. He is an advisor to Roche and Roche Diagnostics, which reimburse his expenses only. T.G.B. has research support/contracts from the National Institutes of Health, State of Arizona, Michael J Fox Foundation, Avid Radiopharmaceuticals, Navida Biopharmaceuticals and Aprinoia Therapeutics. He is an advisory board member with Vivid Genomics and has consultancy work with Roche Diagnostics. A.G.S. conducts multiple industry-funded clinical trials, but all funds go to her academic institution. They have current (within last 12 months) research contracts with Eli Lilly, Novartis, Roche, Janssen, AbbVie, Biogen, NeuroEM, Suven and Merck. She does not receive personal compensations from these organizations. G.D.S. is a consultant for Biogen, Inc. J.M.B. is participating in clinical trials of antidementia drugs for Eli Lilly, Toyama Chemical Company, Merck, Biogen, AbbVie, vTv Therapeutics, Janssen and Roche. He has received research grants from Eli Lilly, Avid Radiopharmaceuticals and Astra Zeneca. He is a consultant for Stage 2 Innovations. L.F. is a consultant for Allergan, Eli Lilly, Avraham Pharmaceuticals, Axon Neuroscience, Axovant, Biogen, Boehringer Ingelheim, Eisai, Functional Neuromodulation, Lundbeck, MerckSharpe & Dohme, Novartis, Pfizer, Pharnext, Roche and Schwabe Pharma. M.B has consulted as an advisory board member for Araclon, Grifols, Lilly, Nutricia, Roche and Servier. She received fees for lectures and funds for research from Araclon, Grifols, Nutricia, Roche and Servier. She has not received personal compensations from these organizations. A.Ruiz has consulted for Grifols and Landsteiner Genmed. He received fees for lectures or funds for research and/or reimbursement of expenses for congresses attendance from Araclon and Grifols. He has not received personal compensations from these organizations. O.P. acts as a consultant for Roche and Biogen, Inc. He is currently participating in clinical trials of antidementia drugs from Novartis, Genentech, Roche and Pharmatrophix. B.T.H. is a consultant for Aztherapy, Biogen, Calico, Ceregene, Genentech, Lilly, Neurophage, Novartis and Takeda, and receives research support from Abbvie, Amgen, Deanli, Fidelity Biosciences, General Electric, Lilly, Merck, Sangamo and Spark therapeutics. BTH owns Novartis stock. H.Hampel serves as Senior Associate Editor for the Journal Alzheimer’s & Dementia; he received lecture fees from Biogen and Roche, research grants from Pfizer, Avid and MSD AVENIR (paid to the institution), travel funding from Functional Neuromodulation, Axovant, Eli Lilly and company, Takeda and Zinfandel, GE Healthcare and Oryzon Genomics, consultancy fees from Jung Diagnostics, Cytox Ltd., Axovant, Anavex, Takeda and Zinfandel, GE Healthcare, Oryzon Genomics and Functional Neuromodulation, and participated in scientific advisory boards of Functional Neuromodulation, Axovant, Eli Lilly and company, Cytox Ltd., GE Healthcare, Takeda and Zinfandel, Oryzon Genomics and Roche Diagnostica. Harald Hampel is a co-inventor on numerous patents relating to biomarker measurement but has received no royalties from these patents. A.A.-C. has consultancies for GSK, Cytokinetics, Biogen Idec, Treeway Inc, Chronos Therapeutics, OrionPharma and Mitsubishi-Tanabe Pharma, and was Chief Investigator for commercial clinical trials run by OrionPharma and Cytokinetics.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figures 1–22 and Supplementary Note
Supplementary Tables
Supplementary Tables 1–31
Rights and permissions
About this article
Cite this article
Kunkle, B.W., Grenier-Boley, B., Sims, R. et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat Genet 51, 414–430 (2019). https://doi.org/10.1038/s41588-019-0358-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-019-0358-2
This article is cited by
-
The role of CD56bright NK cells in neurodegenerative disorders
Journal of Neuroinflammation (2024)
-
Causal relationships between type 1 diabetes mellitus and Alzheimer’s disease and Parkinson’s disease: a bidirectional two-sample Mendelian randomization study
European Journal of Medical Research (2024)
-
Synaptic vesicle glycoprotein 2 A in serum is an ideal biomarker for early diagnosis of Alzheimer’s disease
Alzheimer's Research & Therapy (2024)
-
Chinese English language learners’ vocabulary retention: Investigating the effectiveness of neuro/metacognitive and socio-cultural strategies
BMC Psychology (2024)
-
Unraveling the intercellular communication disruption and key pathways in Alzheimer’s disease: an integrative study of single-nucleus transcriptomes and genetic association
Alzheimer's Research & Therapy (2024)