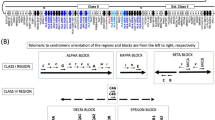
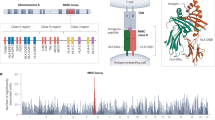
Abstract
To perform detailed fine-mapping of the major-histocompatibility-complex region, we conducted next-generation sequencing (NGS)-based typing of the 33 human leukocyte antigen (HLA) genes in 1,120 individuals of Japanese ancestry, providing a high-resolution allele catalog and linkage-disequilibrium structure of both classical and nonclassical HLA genes. Together with population-specific deep-whole-genome-sequencing data (n = 1,276), we conducted NGS-based HLA, single-nucleotide-variant and indel imputation of large-scale genome-wide-association-study data from 166,190 Japanese individuals. A phenome-wide association study assessing 106 clinical phenotypes identified abundant, significant genotype–phenotype associations across 52 phenotypes. Fine-mapping highlighted multiple association patterns conferring independent risks from classical HLA genes. Region-wide heritability estimates and genetic-correlation network analysis elucidated the polygenic architecture shared across the phenotypes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Code availability
Software and codes used for this study are available from URLs or upon request to the authors.
Data availability
HLA data have been deposited at the National Bioscience Database Center (NBDC) Human Database (research ID: hum0114) as open data without any access restrictions. GWAS data and phenotype data of the BBJ individuals are available at the NBDC Human Database (research ID: hum0014).
References
Dendrou, C. A., Petersen, J., Rossjohn, J. & Fugger, L. HLA variation and disease. Nat. Rev. Immunol. 18, 325–339 (2018).
Okada, Y. et al. HLA-Cw*1202-B*5201-DRB1*1502 haplotype increases risk for ulcerative colitis but reduces risk for Crohn’s disease. Gastroenterology 141, 864–871 (2011).
Okada, Y. et al. Risk for ACPA-positive rheumatoid arthritis is driven by shared HLA amino acid polymorphisms in Asian and European populations. Hum. Mol. Genet. 23, 6916–6926 (2014).
Okada, Y. et al. Construction of a population-specific HLA imputation reference panel and its application to Graves’ disease risk in Japanese. Nat. Genet. 47, 798–802 (2015).
Robinson, J., Soormally, A. R., Hayhurst, J. D. & Marsh, S. G. The IPD-IMGT/HLA Database: new developments in reporting HLA variation. Hum. Immunol. 77, 233–237 (2016).
The MHC sequencing consortium. Complete sequence and gene map of a human major histocompatibility complex. Nature 401, 921–923 (1999).
Horton, R. et al. Gene map of the extended human MHC. Nat. Rev. Genet. 5, 889–899 (2004).
Nejentsev, S. et al. Localization of type 1 diabetes susceptibility to the MHC class I genes HLA-B and HLA-A. Nature 450, 887–892 (2007).
Jia, X. et al. Imputing amino acid polymorphisms in human leukocyte antigens. PLoS One 8, e64683 (2013).
Raychaudhuri, S. et al. Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nat. Genet. 44, 291–296 (2012).
Okada, Y. et al. Fine mapping major histocompatibility complex associations in psoriasis and its clinical subtypes. Am. J. Hum. Genet. 95, 162–172 (2014).
Hirata, J. et al. Variants at HLA-A, HLA-C, and HLA-DQB1 confer risk of psoriasis vulgaris in Japanese. J. Invest. Dermatol. 138, 542–548 (2018).
Hosomichi, K., Shiina, T., Tajima, A. & Inoue, I. The impact of next-generation sequencing technologies on HLA research. J. Hum. Genet. 60, 665–673 (2015).
Zhou, F. et al. Deep sequencing of the MHC region in the Chinese population contributes to studies of complex disease. Nat. Genet. 48, 740–746 (2016).
Robinson, J. et al. Distinguishing functional polymorphism from random variation in the sequences of >10,000 HLA-A, -B and -C alleles. PLoS Genet. 13, e1006862 (2017).
Schofl, G. et al. 2.7 million samples genotyped for HLA by next generation sequencing: lessons learned. BMC Genomics 18, 161 (2017).
Walter, K. et al. The UK10K project identifies rare variants in health and disease. Nature 526, 82–90 (2015).
Okada, Y. et al. Contribution of a nonclassical HLA gene, HLA-DOA, to the risk of rheumatoid arthritis. Am. J. Hum. Genet. 99, 366–374 (2016).
Okushi, Y. et al. Circulating and renal expression of HLA-G prevented chronic renal allograft dysfunction in Japanese recipients. Clin. Exp. Nephrol. 21, 932–940 (2017).
Astle, W. J. et al. The allelic landscape of human blood cell trait variation and links to common complex disease. Cell 167, 1415–1429 (2016).
Nagai, A. et al. Overview of the BioBank Japan Project: study design and profile. J. Epidemiol. 27, S2–S8 (2017).
Hirata, M. et al. Cross-sectional analysis of BioBank Japan clinical data: A large cohort of 200,000 patients with 47 common diseases. J. Epidemiol. 27, S9–S21 (2017).
Pickrell, J. K. et al. Detection and interpretation of shared genetic influences on 42 human traits. Nat. Genet. 48, 709–717 (2016).
Finucane, H. K. et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet. 47, 1228–1235 (2015).
Akiyama, M. et al. Genome-wide association study identifies 112 new loci for body mass index in the Japanese population. Nat. Genet. 49, 1458–1467 (2017).
Kanai, M. et al. Genetic analysis of quantitative traits in the Japanese population links cell types to complex human diseases. Nat. Genet. 50, 390–400 (2018).
Denny, J. C. et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat. Biotechnol. 31, 1102–1110 (2013).
Bush, W. S., Oetjens, M. T. & Crawford, D. C. Unravelling the human genome-phenome relationship using phenome-wide association studies. Nat. Rev. Genet. 17, 129–145 (2016).
Karnes, J. H. et al. Phenome-wide scanning identifies multiple diseases and disease severity phenotypes associated with HLA variants. Sci. Transl. Med. 9, eaai8708 (2017).
Okada, Y. et al. Deep whole-genome sequencing reveals recent selection signatures linked to evolution and disease risk of Japanese. Nat. Commun. 9, 1631 (2018).
Hosomichi, K. et al. Phase-defined complete sequencing of the HLA genes by next-generation sequencing. BMC Genomics 14, 355 (2013).
Hosomichi, K., Mitsunaga, S., Nagasaki, H. & Inoue, I. A bead-based normalization for uniform sequencing depth (BeNUS) protocol for multi-samples sequencing exemplified by HLA-B. BMC Genomics 15, 645 (2014).
Yang, K. L. et al. New allele name of some HLA-DRB1*1401: HLA-DRB1*1454. Int. J. Immunogenet. 36, 119–120 (2009).
Morizane, A. et al. MHC matching improves engraftment of iPSC-derived neurons in non-human primates. Nat. Commun. 8, 385 (2017).
van der Maaten, L. & Hilton, G. Visualizing high-dimensional data using t-SNE. J. Mach. Learn. Res. 9, 2579–2605 (2008).
van der Maaten, L. Visualizing data using t-SNE. J. Mach. Learn. Res. 15, 3221–3245 (2014).
Ester, M., Kriegel, H. P., Sander, J. & Xu, X. A density-based algorithm for discovering clusters in large spatial databases with noise. in KDD ‘96 Proc. Second Int. Conf. Knowl. Discov. Data Min., 226–231 (AAAI Press, Palo Alto, CA, USA, 1996).
Bauer, D. C., Zadoorian, A., Wilson, L. O. W. & Thorne, N. P. Evaluation of computational programs to predict HLA genotypes from genomic sequencing data. Brief. Bioinform. 19, 179–187 (2018).
Kanai, M., Tanaka, T. & Okada, Y. Empirical estimation of genome-wide significance thresholds based on the 1000 Genomes Project data set. J. Hum. Genet. 61, 861–866 (2016).
Nishida, N. et al. Understanding of HLA-conferred susceptibility to chronic hepatitis B infection requires HLA genotyping-based association analysis. Sci. Rep. 6, 24767 (2016).
Okada, Y. et al. Identification of nine novel loci associated with white blood cell subtypes in a Japanese population. PLoS Genet. 7, e1002067 (2011).
Ikeshita, S., Miyatake, Y., Otsuka, N. & Kasahara, M. MICA/B expression in macrophage foam cells infiltrating atherosclerotic plaques. Exp. Mol. Pathol. 97, 171–175 (2014).
Hu, X. et al. Additive and interaction effects at three amino acid positions in HLA-DQ and HLA-DR molecules drive type 1 diabetes risk. Nat. Genet. 47, 898–905 (2015).
Roe, D. et al. Revealing complete complex KIR haplotypes phased by long-read sequencing technology. Genes Immun. 18, 127–134 (2017).
See, P. et al. Mapping the human DC lineage through the integration of high-dimensional techniques. Science 356, eaag3009 (2017).
Takeuchi, Y. et al. Clinical response to PD-1 blockade correlates with a sub-fraction of peripheral central memory CD4+ T cells in patients with malignant melanoma. Int. Immunol. 30, 13–22 (2018).
Platzer, A. Visualization of SNPs with t-SNE. PLoS One 8, e56883 (2013).
Shi, H., Mancuso, N., Spendlove, S. & Pasaniuc, B. Local genetic correlation gives insights into the shared genetic architecture of complex traits. Am. J. Hum. Genet. 101, 737–751 (2017).
Szolek, A. et al. OptiType: precision HLA typing from next-generation sequencing data. Bioinformatics 30, 3310–3316 (2014).
Shukla, S. A. et al. Comprehensive analysis of cancer-associated somatic mutations in class I HLA genes. Nat. Biotechnol. 33, 1152–1158 (2015).
Kawaguchi, S. et al. HLA-HD: an accurate HLA typing algorithm for next-generation sequencing data. Hum. Mutat. 38, 788–797 (2017).
Lee, H. & Kingsford, C. Kourami: graph-guided assembly for novel human leukocyte antigen allele discovery. Genome. Biol. 19, 16 (2018).
Nothnagel, M. & Rohde, K. The effect of single-nucleotide polymorphism marker selection on patterns of haplotype blocks and haplotype frequency estimates. Am. J. Hum. Genet. 77, 988–998 (2005).
Okada, Y. eLD: entropy-based linkage disequilibrium index between multi-allelic sites. Hum. Genome Var. 5, 29 (2018).
Okada, Y. et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature 506, 376–381 (2014).
Karnes, J. H. et al. Comparison of HLA allelic imputation programs. PLoS One 12, e0172444 (2017).
Lenz, T. L. et al. Widespread non-additive and interaction effects within HLA loci modulate the risk of autoimmune diseases. Nat. Genet. 47, 1085–1090 (2015).
Yang, J., Lee, S. H., Goddard, M. E. & Visscher, P. M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88, 76–82 (2011).
Lee, S. H., Wray, N. R., Goddard, M. E. & Visscher, P. M. Estimating missing heritability for disease from genome-wide association studies. Am. J. Hum. Genet. 88, 294–305 (2011).
Speed, D. et al. Reevaluation of SNP heritability in complex human traits. Nat. Genet. 49, 986–992 (2017).
Hinks, A. et al. Fine-mapping the MHC locus in juvenile idiopathic arthritis (JIA) reveals genetic heterogeneity corresponding to distinct adult inflammatory arthritic diseases. Ann. Rheum. Dis. 76, 765–772 (2017).
Acknowledgements
We thank T. Aoi for kind support of the study. This research was supported by the Tailor-Made Medical Treatment program (the BioBank Japan Project) of the Ministry of Education, Culture, Sports, Science and Technology (MEXT), the Japan Agency for Medical Research and Development (AMED), MEXT KAKENHI (221S0002), Bioinformatics Initiative of Osaka University Graduate School of Medicine, and Integrated Frontier Research for Medical Science Division, Institute for Open and Transdisciplinary Research Initiatives, Osaka University. Y.O. was supported by the Japan Society for the Promotion of Science (JSPS) KAKENHI (15H05670, 15H05911 and 15K14429), AMED (18gm6010001h0003 and 18ek0410041h0002), Takeda Science Foundation, the Uehara Memorial Foundation, the Naito Foundation, Daiichi Sankyo Foundation of Life Science, Senri Life Science Foundation and Suzuken Memorial Foundation. K.H. was supported by JSPS KAKENHI grant no. P16H06502 ‘Neo-self’. J.H. is an employee of Teijin Pharma Limited. Computations were partially performed on the NIG supercomputer at ROIS National Institute of Genetics.
Author information
Authors and Affiliations
Contributions
Y.O. supervised the study. J.H., K.H., Y.K. and Y.O. wrote the manuscript. J.H., K.H., S.S., M. Kanai, K.I., K.S., M.A., T.K., K.O., T.M., K.Y., Y.K. and Y.O. conducted data analysis. M.H., K.M., M. Kubo and Y.K. provided data. Y.O., K.M. and M. Kubo collected samples. Y.O., K.H., H.N., Y.M. and I.I. conducted experiments.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1, 2 and 4–7, and Supplementary Figures 1–3
Supplementary Tables
Supplementary Tables 3 and 8
Rights and permissions
About this article
Cite this article
Hirata, J., Hosomichi, K., Sakaue, S. et al. Genetic and phenotypic landscape of the major histocompatibilty complex region in the Japanese population. Nat Genet 51, 470–480 (2019). https://doi.org/10.1038/s41588-018-0336-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-018-0336-0
This article is cited by
-
Tutorial: a statistical genetics guide to identifying HLA alleles driving complex disease
Nature Protocols (2023)
-
Contribution of an Asian-prevalent HLA haplotype to the risk of HBV-related hepatocellular carcinoma
Scientific Reports (2023)
-
Pan-cancer and cross-population genome-wide association studies dissect shared genetic backgrounds underlying carcinogenesis
Nature Communications (2023)
-
Multi-ancestry meta-analysis and fine-mapping in Alzheimer’s disease
Molecular Psychiatry (2023)
-
Genome-Wide Association Study of Intracranial Artery Stenosis Followed by Phenome-Wide Association Study
Translational Stroke Research (2023)