Abstract
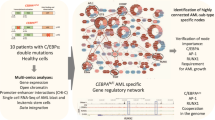
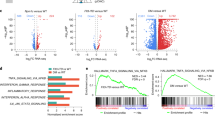
Acute myeloid leukemia (AML) is a heterogeneous disease caused by a variety of alterations in transcription factors, epigenetic regulators and signaling molecules. To determine how different mutant regulators establish AML subtype–specific transcriptional networks, we performed a comprehensive global analysis of cis-regulatory element activity and interaction, transcription factor occupancy and gene expression patterns in purified leukemic blast cells. Here, we focused on specific subgroups of subjects carrying mutations in genes encoding transcription factors (RUNX1, CEBPα), signaling molecules (FTL3-ITD, RAS) and the nuclear protein NPM1). Integrated analysis of these data demonstrates that each mutant regulator establishes a specific transcriptional and signaling network unrelated to that seen in normal cells, sustaining the expression of unique sets of genes required for AML growth and maintenance.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
Raw data have been deposited at GEO under accession number GSE108316. Processed data are available from our data server (http://bioinformatics-bham.co.uk/tfinaml/).
References
Cancer Genome Atlas Research, N. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 368, 2059–2074 (2013).
Papaemmanuil, E. et al. Genomic classification and prognosis in acute myeloid leukemia. N. Engl. J. Med. 374, 2209–2221 (2016).
Bonifer, C. & Cockerill, P. N. Chromatin structure profiling identifies crucial regulators of tumor maintenance. Trends Cancer 1, 157–160 (2015).
Rosenbauer, F. & Tenen, D. G. Transcription factors in myeloid development: balancing differentiation with transformation. Nat. Rev. Immunol. 7, 105–117 (2007).
Cockerill, P. N. Receptor signaling directs global recruitment of pre-existing transcription factors to inducible elements. Yale J. Biol. Med. 89, 591–596 (2016).
Ward, A. F., Braun, B. S. & Shannon, K. M. Targeting oncogenic Ras signaling in hematologic malignancies. Blood 120, 3397–3406 (2012).
Parikh, C., Subrahmanyam, R. & Ren, R. Oncogenic NRAS rapidly and efficiently induces CMML- and AML-like diseases in mice. Blood 108, 2349–2357 (2006).
Masson, K. & Ronnstrand, L. Oncogenic signaling from the hematopoietic growth factor receptors c-Kit and Flt3. Cell Signal. 21, 1717–1726 (2009).
Gerloff, D. et al. NF-kappaB/STAT5/miR-155 network targets PU.1 in FLT3-ITD-driven acute myeloid leukemia. Leukemia 29, 535–547 (2015).
Corces-Zimmerman, M. R., Hong, W. J., Weissman, I. L., Medeiros, B. C. & Majeti, R. Preleukemic mutations in human acute myeloid leukemia affect epigenetic regulators and persist in remission. Proc. Natl Acad. Sci. USA 111, 2548–2553 (2014).
Shlush, L. I. et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature 506, 328–333 (2014).
Di Croce, L. & Helin, K. Transcriptional regulation by Polycomb group proteins. Nat. Struct. Mol. Biol. 20, 1147–1155 (2013).
Broske, A. M. et al. DNA methylation protects hematopoietic stem cell multipotency from myeloerythroid restriction. Nat. Genet. 41, 1207–1215 (2009).
Bonifer, C. & Bowen, D. T. Epigenetic mechanisms regulating normal and malignant haematopoiesis: new therapeutic targets for clinical medicine. Expert Rev. Mol. Med. 12, e6 (2010).
Challen, G. A. et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat. Genet. 44, 23–31 (2012).
Ko, M. et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature 468, 839–843 (2010).
Shih, A. H., Abdel-Wahab, O., Patel, J. P. & Levine, R. L. The role of mutations in epigenetic regulators in myeloid malignancies. Nat. Rev. Cancer 12, 599–612 (2012).
Goode, D. K. et al. Dynamic gene regulatory networks drive hematopoietic specification and differentiation. Dev. Cell 36, 572–587 (2016).
Obier, N. & Bonifer, C. Chromatin programming by developmentally regulated transcription factors: lessons from the study of haematopoietic stem cell specification and differentiation. FEBS Lett. 590, 4105–4115 (2016).
Wilson, N. K. et al. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell 7, 532–544 (2010).
Rubin, A. J. et al. Lineage-specific dynamic and pre-established enhancer-promoter contacts cooperate in terminal differentiation. Nat. Genet. 49, 1522–1528 (2017).
Ptasinska, A. et al. Identification of a dynamic core transcriptional network in t(8;21) AML that regulates differentiation block and self-renewal. Cell Rep. 8, 1974–1988 (2014).
Loke, J. et al. RUNX1-ETO and RUNX1-EVI1 differentially reprogram the chromatin landscape in t(8;21) and t(3;21) AML. Cell Rep. 19, 1654–1668 (2017).
Cauchy, P. et al. Chronic FLT3-ITD signaling in acute myeloid leukemia is connected to a specific chromatin signature. Cell Rep. 12, 821–836 (2015).
Corces, M. R. et al. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat. Genet. 48, 1193–1203 (2016).
McKeown, M. R. et al. Superenhancer analysis defines novel epigenomic subtypes of non-APL AML, including an RARalpha dependency targetable by SY-1425, a potent and selective RARalpha agonist. Cancer Discov. 7, 1136–1153 (2017).
Martens, J. H. et al. ERG and FLI1 binding sites demarcate targets for aberrant epigenetic regulation by AML1-ETO in acute myeloid leukemia. Blood 120, 4038–4048 (2012).
Pulikkan, J. A., Tenen, D. G. & Behre, G. C/EBPalpha deregulation as a paradigm for leukemogenesis. Leukemia 31, 2279–2285 (2017).
Piper, J. et al. Wellington: a novel method for the accurate identification of digital genomic footprints from DNase-seq data. Nucleic Acids Res. 41, e201 (2013).
Ptasinska, A. et al. Depletion of RUNX1/ETO in t(8;21) AML cells leads to genome-wide changes in chromatin structure and transcription factor binding. Leukemia 26, 1829–1841 (2012).
Mandoli, A. et al. CBFB-MYH11/RUNX1 together with a compendium of hematopoietic regulators, chromatin modifiers and basal transcription factors occupies self-renewal genes in inv(16) acute myeloid leukemia. Leukemia 28, 770–778 (2014).
Dunne, J. et al. AML1/ETO proteins control POU4F1/BRN3A expression and function in t(8;21) acute myeloid leukemia. Cancer Res. 70, 3985–3995 (2010).
Dekker, J., Rippe, K., Dekker, M. & Kleckner, N. Capturing chromosome conformation. Science 295, 1306–1311 (2002).
Mifsud, B. et al. Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat. Genet. 47, 598–606 (2015).
Chasman, D. & Roy, S. Inference of cell type specific regulatory networks on mammalian lineages. Curr. Opin. Syst. Biol. 2, 130–139 (2017).
Neph, S. et al. Circuitry and dynamics of human transcription factor regulatory networks. Cell 150, 1274–1286 (2012).
Lin, S. et al. A FOXO1-induced oncogenic network defines the AML1-ETO preleukemic program. Blood 130, 1213–1222 (2017).
O’Connor, C. et al. Nfix expression critically modulates early B lymphopoiesis and myelopoiesis. PLoS One 10, e0120102 (2015).
Somerville, T. D. et al. Frequent derepression of the mesenchymal transcription factor gene FOXC1 in acute myeloid leukemia. Cancer Cell 28, 329–342 (2015).
Olive, M. et al. A dominant negative to activation protein-1 (AP1) that abolishes DNA binding and inhibits oncogenesis. J. Biol. Chem. 272, 18586–18594 (1997).
Obier, N. et al. Cooperative binding of AP-1 and TEAD4 modulates the balance between vascular smooth muscle and hemogenic cell fate. Development 143, 4324–4340 (2016).
Verhaak, R. G. W. et al. Prediction of molecular subtypes in acute myeloid leukemia based on gene expression profiling. Haematologica 94, 131–134 (2009).
Figueroa, M. E. et al. DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer Cell 17, 13–27 (2010).
Lin, S., Mulloy, J. C. & Goyama, S. RUNX1-ETO leukemia. Adv. Exp. Med. Biol. 962, 151–173 (2017).
Goyama, S. et al. UBASH3B/Sts-1-CBL axis regulates myeloid proliferation in human preleukemia induced by AML1-ETO. Leukemia 30, 728–739 (2016).
Sun, X. J. et al. A stable transcription factor complex nucleated by oligomeric AML1-ETO controls leukaemogenesis. Nature 500, 93–97 (2013).
Essig, A., Duque-Afonso, J., Schwemmers, S., Pahl, H. L. & Lubbert, M. The AML1/ETO target gene LAT2 interferes with differentiation of normal hematopoietic precursor cells. Leuk. Res. 38, 340–345 (2014).
Trop-Steinberg, S. & Azar, Y. AP-1 expression and its clinical relevance in immune disorders and cancer. Am. J. Med. Sci. 353, 474–483 (2017).
Martinez-Soria, N. et al. The Oncogenic Transcription Factor RUNX1/ETO Corrupts Cell Cycle Regulation to Drive Leukemic Transformation. Cancer Cell 34, 626–642.e8 (2018).
Kesarwani, M. et al. Targeting c-FOS and DUSP1 abrogates intrinsic resistance to tyrosine-kinase inhibitor therapy in BCR-ABL-induced leukemia. Nat. Med. 23, 472–482 (2017).
Faber, Z. J. et al. The genomic landscape of core-binding factor acute myeloid leukemias. Nat. Genet. 48, 1551–1556 (2016).
Levis, M. et al. Results from a randomized trial of salvage chemotherapy followed by lestaurtinib for patients with FLT3 mutant AML in first relapse. Blood 117, 3294–3301 (2011).
DePristo, M. A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498 (2011).
van Gosliga, D. et al. Establishing long-term cultures with self-renewing acute myeloid leukemia stem/progenitor cells. Exp. Hematol. 35, 1538–1549 (2007).
Weber, K., Bartsch, U., Stocking, C. & Fehse, B. A multicolor panel of novel lentiviral “gene ontology” (LeGO) vectors for functional gene analysis. Mol. Ther. 16, 698–706 (2008).
Mostoslavsky, G. et al. Efficiency of transduction of highly purified murine hematopoietic stem cells by lentiviral and oncoretroviral vectors under conditions of minimal in vitro manipulation. Mol. Ther. 11, 932–940 (2005).
Bert, A. G., Johnson, B. V., Baxter, E. W. & Cockerill, P. N. A modular enhancer is differentially regulated by GATA and NFAT elements that direct different tissue-specific patterns of nucleosome positioning and inducible chromatin remodeling. Mol. Cell. Biol. 27, 2870–2885 (2007).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Trapnell, C. et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31, 46–53 (2013).
Wingett, S. et al. HiCUP: pipeline for mapping and processing Hi-Cdata. F1000Res. 4, 1310 (2015).
Mifsud, B. et al. GOTHiC, a probabilistic model to resolve complex biases and to identify real interactions in Hi-C data. PLoS One 12, e0174744 (2017).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Zhang, H. M. et al. AnimalTFDB: a comprehensive animal transcription factor database. Nucleic Acids Res. 40, D144–D149 (2012).
Acknowledgements
This research was funded by a program grant from Bloodwise (15001) to C.B. and P.N.C., as well as studentship awards from Cancer Research UK and Bloodwise to A.Pickin and N.G., respectively, a Kay Kendall Clinical Training Fellowship for J.L. and a MRC/Leuka Clinical Training Fellowship for S.P.
Author information
Authors and Affiliations
Contributions
M.R.I., D.J.L.C., S.P., A.Ptasinska, H.B., A.Pickin, L.N.G., J.C.L., P.S.C., J.Z.-C. and S.R.J. performed experiments and generated data; R.D., M.R., S.J.R., M.J.G., P.J. and A.U. provided subject samples; S.C., A.B. and P.N.C. conducted mutation analysis; S.A.A. and P.C. analyzed data; O.H. supervised transplantation experiments and helped edit the manuscript; C.B. and P.N.C. conceived and directed the study and C.B. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–10, Supplementary Tables 1–3 and Supplementary Note
Supplementary Data 1
Summary of all AML mutation data
Supplementary Data 2
Up- and downregulated genes associated with mutation groups
Supplementary Data 3
Number of RNA-seq differentially expressed genes for Fig. SN1 and Supplementary Fig. 4c
Supplementary Data 4
AML subtype-specific transcription factor gene expression
Supplementary Data 6
Gene lists and GO terms for Supplementary Figs. 5 and 7
Supplementary Data 7
Statistical data fmr validations
Rights and permissions
About this article
Cite this article
Assi, S.A., Imperato, M.R., Coleman, D.J.L. et al. Subtype-specific regulatory network rewiring in acute myeloid leukemia. Nat Genet 51, 151–162 (2019). https://doi.org/10.1038/s41588-018-0270-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-018-0270-1
This article is cited by
-
Data-driven modeling of core gene regulatory network underlying leukemogenesis in IDH mutant AML
npj Systems Biology and Applications (2024)
-
Leukemic stem cells activate lineage inappropriate signalling pathways to promote their growth
Nature Communications (2024)
-
Gene regulation in t(6;9) DEK::NUP214 Acute Myeloid Leukemia resembles that of FLT3-ITD/NPM1 Acute Myeloid Leukemia but with an altered HOX/MEIS axis
Leukemia (2024)
-
Regulation of HOX gene expression in AML
Blood Cancer Journal (2024)
-
LRRC1 knockdown downregulates MACF1 to inhibit the malignant progression of acute myeloid leukemia by inactivating β-catenin/c-Myc signaling
Journal of Molecular Histology (2024)