Abstract
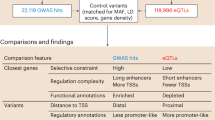
We introduce new statistical methods for analyzing genomic data sets that measure many effects in many conditions (for example, gene expression changes under many treatments). These new methods improve on existing methods by allowing for arbitrary correlations in effect sizes among conditions. This flexible approach increases power, improves effect estimates and allows for more quantitative assessments of effect-size heterogeneity compared to simple shared or condition-specific assessments. We illustrate these features through an analysis of locally acting variants associated with gene expression (cis expression quantitative trait loci (eQTLs)) in 44 human tissues. Our analysis identifies more eQTLs than existing approaches, consistent with improved power. We show that although genetic effects on expression are extensively shared among tissues, effect sizes can still vary greatly among tissues. Some shared eQTLs show stronger effects in subsets of biologically related tissues (for example, brain-related tissues), or in only one tissue (for example, testis). Our methods are widely applicable, computationally tractable for many conditions and available online.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The GTEx study data are available through dbGaP under accession phs000424.v6.p1. The GTEx summary statistics used in the mash analysis have been deposited in Zenodo (https://doi.org/10.5281/zenodo.1296399).
References
Blischak, J. D., Tailleux, L., Mitrano, A., Barreiro, L. B. & Gilad, Y. Mycobacterial infection induces a specific human innate immune response. Sci. Rep. 5, 16882 (2015).
Ferguson, J. P., Cho, J. H. & Zhao, H. A new approach for the joint analysis of multiple ChIP-Seq libraries with application to histone modification. Stat. Appl. Genet. Mol. Biol. 11, https://doi.org/10.1515/1544-6115.1660 (2012).
Pickrell, J., Berisa, T., Ségurel, L., Tung, J. Y. & Hinds, D. Detection and interpretation of shared genetic influences on 40 human traits. Nat. Genet. 48, 709–717 (2016).
Dimas, A. S. et al. Common regulatory variation impacts gene expression in a cell type-dependent manner. Science 325, 1246–1250 (2009).
Flutre, T., Wen, X., Pritchard, J. & Stephens, M. A statistical framework for joint eQTL analysis in multiple tissues. PLoS Genet. 9, e1003486 (2013).
Li, G., Shabalin, A. A., Rusyn, I., Wright, F. A. & Nobel, A. B. An Empirical Bayes approach for multiple tissue eQTL Analysis. Biostatistics 19, 391–406 (2017).
Petretto, E. et al. New insights into the genetic control of gene expression using a Bayesian multi-tissue approach. PLoS Comput. Biol. 6, e1000737 (2010).
Wen, X. & Stephens, M. Using linear predictors to impute allele frequencies from summary of pooled genotype data. Ann. Appl. Stat. 4, 1158–1182 (2010).
Han, B. & Eskin, E. Random-effects model aimed at discovering associations in meta-analysis of genome-wide association studies. Am. J. Hum. Genet. 88, 586–598 (2011).
Stephens, M. Unified framework for association analysis with multiple related phenotypes. PLoS One 8, e65245 (2013).
Sul, J. H., Han, B., Ye, C., Choi, T. & Eskin, E. Effectively identifying eQTLs from multiple tissues by combining mixed model and meta-analytic approaches. PLoS Genet. 9, e1003491 (2013).
Wei, Y., Tenzen, T. & Ji, H. Joint analysis of differential gene expression in multiple studies using correlation motifs. Biostatistics 16, 31–46 (2015).
Zhou, X. & Stephens, M. Efficient multivariate linear mixed model algorithms for genome-wide association studies. Nat. Methods 11, 407–409 (2014).
Han, B. & Eskin, E. Interpreting meta-analyses of genome-wide association studies. PLoS Genet. 8, e1002555 (2012).
Lebrec, J. J., Stijnen, T. & van Houwelingen, H. C. Dealing with heterogeneity between cohorts in genomewide SNP association studies. Stat. Appl. Genet. Mol. Biol. 9, https://doi.org/10.2202/1544-6115.1503 (2010).
Stephens, M. False discovery rates: a new deal. Biostatistics 18, 275–294 (2017).
GTEx Consortium. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348, 648–660 (2015).
Nicolae, D. L. et al. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 6, e1000888 (2010).
Shabalin, A. A. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics 28, 1353–1358 (2012).
Price, A. L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006).
Engelhardt, B. E. & Stephens, M. Analysis of population structure: a unifying framework and novel methods based on sparse factor analysis. PLoS Genet. 6, e1001117 (2010).
Tversky, A. & Kahneman, D. Judgment under uncertainty: heuristics and biases. Science 185, 1124–1131 (1974).
Wen, X. & Stephens, M. Bayesian methods for genetic association analysis with heterogeneous subgroups: from meta-analyses to gene-environment interactions. Ann. Appl. Stat. 8, 176–203 (2014).
Servin, B. & Stephens, M. Imputation-based analysis of association studies: candidate regions and quantitative traits. PLoS Genet. 3, e114 (2007).
Veyrieras, J.-B. et al. High-resolution mapping of expression-QTLs yields insight into human gene regulation. PLoS Genet. 4, e1000214 (2008).
Hormozdiari, F., Kostem, E., Kang, E. Y., Pasaniuc, B. & Eskin, E. Identifying causal variants at loci with multiple signals of association. Genetics 198, 497–508 (2014).
Kichaev, G. et al. Integrating functional data to prioritize causal variants in statistical fine-mapping studies. PLoS Genet. 10, e1004722 (2014).
Chen, W. et al. Fine mapping causal variants with an approximate Bayesian method using marginal test statistics. Genetics 200, 719–736 (2015).
Moyerbrailean, G. A. et al. Which genetic variants in DNase-seq footprints are more likely to alter binding? PLoS Genet. 12, e1005875 (2016).
Giambartolomei, C. et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 10, e1004383 (2014).
Fortune, M. D. et al. Statistical colocalization of genetic risk variants for related autoimmune diseases in the context of common controls. Nat. Genet. 47, 839–846 (2015).
Nica, A. C. et al. Candidate causal regulatory effects by integration of expression QTLs with complex trait genetic associations. PLoS Genet. 6, e1000895 (2010).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 57, 289–300 (1995).
Storey, J. D. The positive false discovery rate: a Bayesian interpretation and the q-value. Ann. Stat. 31, 2013–2035 (2003).
Bovy, J., Hogg, D. W. & Roweis, S. T. Extreme Deconvolution: inferring complete distribution functions from noisy, heterogeneous and incomplete observations. Ann. Appl. Stat. 5, 1657–1677 (2011).
Larribe, F. & Fearnhead, P. Composite likelihood methods in statistical genetics. Stat. Sin. 21, 43–69 (2011).
Dempster, A. P., Laird, N. M. & Rubin, D. B. Maximum likelihood from incomplete data via the EM algorithm. J. R. Stat. Soc. Series B Stat. Methodol. 39, 1–38 (1977).
Varadhan, R. & Roland, C. Simple and globally convergent methods for accelerating the convergence of any EM algorithm. Scand. J. Stat. 35, 335–353 (2008).
Efron, B. Microarrays empirical Bayes and the two-groups model. Stat. Sci. 23, 1–22 (2008).
Acknowledgements
This work was supported by National Institutes of Health (NIH) grants MH090951 and HG02585 to M.S., and by a grant from the Gordon and Betty Moore Foundation (GBMF 4559) to M.S. S.M.U. was supported by NIH grant T32HD007009. Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the NIH. Additional funds were provided by the National Cancer Institute (NCI), National Human Genome Research Institute, National Heart, Lung, and Blood Institute, National Institute on Drug Abuse, National Institute of Mental Health, and National Institute of Neurological Disorders and Stroke. Donors were enrolled at Biospecimen Source Sites funded by NCI/SAIC-Frederick (SAIC-F) subcontracts to the National Disease Research Interchange (10XS170), Roswell Park Cancer Institute (10XS171), and Science Care (X10S172). The Laboratory, Data Analysis, and Coordinating Center was funded through a contract (HHSN268201000029C) to the Broad Institute. Biorepository operations were funded through an SAIC-F subcontract to Van Andel Institute (10ST1035). Additional data repository and project management were provided by SAIC-F (HHSN261200800001E). The Brain Bank was supported by a supplement to University of Miami grants DA006227 and DA033684 and to contract N01MH000028. Statistical methods development grants were made to the University of Geneva (MH090941 and MH101814), the University of Chicago (MH090951, MH090937, MH101820 and MH101825), the University of North Carolina at Chapel Hill (MH090936 and MH101819), Harvard University (MH090948), Stanford University (MH101782), Washington University in St. Louis (MH101810) and the University of Pennsylvania (MH101822). The data used for the analyses described in this manuscript were obtained from the GTEx Portal on 17 October 2015.
Author information
Authors and Affiliations
Contributions
S.M.U. and M.S. conceived of the project and developed the statistical methods. S.M.U. implemented the comparisons with simulated data. S.M.U. and G.W. performed the analyses of the GTEx data and additional analyses. S.M.U., G.W. and M.S. implemented the software, with contributions from P.C. S.M.U. and M.S. wrote the manuscript, with input from G.W. and P.C. P.C. and G.W. prepared the online code and data resources.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–9, Supplementary Tables 1–4 and Supplementary Note
Rights and permissions
About this article
Cite this article
Urbut, S.M., Wang, G., Carbonetto, P. et al. Flexible statistical methods for estimating and testing effects in genomic studies with multiple conditions. Nat Genet 51, 187–195 (2019). https://doi.org/10.1038/s41588-018-0268-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-018-0268-8
This article is cited by
-
Expression- and splicing-based multi-tissue transcriptome-wide association studies identified multiple genes for breast cancer by estrogen-receptor status
Breast Cancer Research (2024)
-
Identifying regulatory loci across 38 lung cell types
Nature Genetics (2024)
-
A landscape of gene expression regulation for synovium in arthritis
Nature Communications (2024)
-
Epigenetic variation impacts individual differences in the transcriptional response to influenza infection
Nature Genetics (2024)
-
Cell-type-specific and disease-associated expression quantitative trait loci in the human lung
Nature Genetics (2024)