Abstract
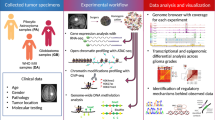
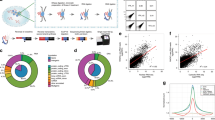
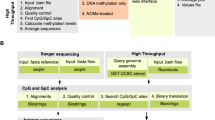
The human genome encodes a variety of poorly understood RNA species that remain challenging to identify using existing genomic tools. We developed chromatin run-on and sequencing (ChRO-seq) to map the location of RNA polymerase for almost any input sample, including samples with degraded RNA that are intractable to RNA sequencing. We used ChRO-seq to map nascent transcription in primary human glioblastoma (GBM) brain tumors. Enhancers identified in primary GBMs resemble open chromatin in the normal human brain. Rare enhancers that are activated in malignant tissue drive regulatory programs similar to the developing nervous system. We identified enhancers that regulate groups of genes that are characteristic of each known GBM subtype and transcription factors that drive them. Finally we discovered a core group of transcription factors that control the expression of genes associated with clinical outcomes. This study characterizes the transcriptional landscape of GBM and introduces ChRO-seq as a method to map regulatory programs that contribute to complex diseases.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
All ChRO-seq and leChRO-seq data can be downloaded from the database of Genotypes and Phenotypes (dbGaP) under accession phs001646.v1.p1. Data collected from Jurkat T cells are available in the Gene Expression Omnibus under accession GSE117832.
References
Cheng, J. et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science 308, 1149–1154 (2005).
Kim, T.-K. et al. Widespread transcription at neuronal activity-regulated enhancers. Nature 465, 182–187 (2010).
Ulitsky, I. & Bartel, D. P. LincRNAs: genomics, evolution, and mechanisms. Cell 154, 26–46 (2013).
Quinodoz, S. & Guttman, M. Long noncoding RNAs: an emerging link between gene regulation and nuclear organization. Trends. Cell Biol. 24, 651–663 (2014).
De Santa, F. et al. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PloS Biol. 8, e1000384 (2010).
Preker, P. et al. RNA exosome depletion reveals transcription upstream of active human promoters. Science 322, 1851–1854 (2008).
Andersson, R. et al. Nuclear stability and transcriptional directionality separate functionally distinct RNA species. Nat. Commun. 5, 5336 (2014).
Core, L. J. et al. Defining the status of RNA polymerase at promoters. Cell Rep. 2, 1025–1035 (2012).
Boyle, A. P. et al. High-resolution mapping and characterization of open chromatin across the genome. Cell 132, 311–322 (2008).
Buenrostro, J. D., Giresi, P. G., Zaba, L. C., Chang, H. Y. & Greenleaf, W. J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013).
Core, L. J., Waterfall, J. J. & Lis, J. T. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322, 1845–1848 (2008).
Churchman, L. S. & Weissman, J. S. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature 469, 368–373 (2011).
Kwak, H., Fuda, N. J., Core, L. J. & Lis, J. T. Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science 339, 950–953 (2013).
Mayer, A. et al. Native elongating transcript sequencing reveals human transcriptional activity at nucleotide resolution. Cell 161, 541–554 (2015).
Nojima, T. et al. Mammalian NET-Seq reveals genome-wide nascent transcription coupled to RNA processing. Cell 161, 526–540 (2015).
Schwalb, B. et al. TT-seq maps the human transient transcriptome. Science 352, 1225–1228 (2016).
Core, L. J. et al. Analysis of nascent RNA identifies a unified architecture of initiation regions at mammalian promoters and enhancers. Nat. Genet. 46, 1311–1320 (2014).
Scruggs, B. S. et al. Bidirectional transcription arises from two distinct hubs of transcription factor binding and active chromatin. Mol. Cell 58, 1101–1112 (2015).
Danko, C. G. et al. Identification of active transcriptional regulatory elements from GRO-seq data. Nat. Methods 12, 433–438 (2015).
Azofeifa, J. G. & Dowell, R. D. A generative model for the behavior of RNA polymerase. Bioinformatics 33, 227–234 (2016).
Andersson, R. et al. An atlas of active enhancers across human cell types and tissues. Nature 507, 455–461 (2014).
Bradner, J. E., Hnisz, D. & Young, R. A. Transcriptional addiction in cancer. Cell 168, 629–643 (2017).
Parsons, D. W. et al. An integrated genomic analysis of human glioblastoma multiforme. Science 321, 1807–1812 (2008).
Brennan, C. W. et al. The somatic genomic landscape of glioblastoma. Cell 155, 462–477 (2013).
Mohan, M., Lin, C., Guest, E. & Shilatifard, A. Licensed to elongate: a molecular mechanism for MLL-based leukaemogenesis. Nat. Rev. Cancer 10, 721–728 (2010).
Wuarin, J. & Schibler, U. Physical isolation of nascent RNA chains transcribed by RNA polymerase II: evidence for cotranscriptional splicing. Mol. Cell. Biol. 14, 7219–7225 (1994).
Mahat, D. B. et al. Base-pair-resolution genome-wide mapping of active RNA polymerases using precision nuclear run-on (PRO-seq). Nat. Protoc. 11, 1455–1476 (2016).
Khodor, Y. L. et al. Nascent-seq indicates widespread cotranscriptional pre-mRNA splicing in Drosophila. Genes Dev. 25, 2502–2512 (2011).
Menet, J. S., Rodriguez, J., Abruzzi, K. C. & Rosbash, M. Nascent-Seq reveals novel features of mouse circadian transcriptional regulation. eLife 1, e00011 (2012).
Cai, H. & Luse, D. S. Transcription initiation by RNA polymerase II in vitro. Properties of preinitiation, initiation, and elongation complexes. J. Biol. Chem. 262, 298–304 (1987).
Choder, M. & Aloni, Y. RNA polymerase II allows unwinding and rewinding of the DNA and thus maintains a constant length of the transcription bubble. J. Biol. Chem. 263, 12994–13002 (1988).
Verhaak, R. G. W. et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 17, 98–110 (2010).
Patel, A. P. et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344, 1396–1401 (2014).
Wang, Q. et al. Tumor evolution of glioma-intrinsic gene expression subtypes associates with immunological changes in the microenvironment. Cancer Cell 32, 42–56.e6 (2017).
Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455, 1061–1068 (2008).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Liu, S. J. et al. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science 355, eaah7111 (2016).
Xi, Z. et al. Overexpression of miR-29a reduces the oncogenic properties of glioblastoma stem cells by downregulating Quaking gene isoform 6. Oncotarget 8, 24949–24963 (2017).
Ma, Y. et al. PVT1 affects growth of glioma microvascular endothelial cells by negatively regulating miR-186. Tumour Biol. 39, 1010428317694326 (2017).
Zhao, D. et al. Heat shock protein 47 regulated by miR-29a to enhance glioma tumor growth and invasion. J. Neurooncol. 118, 39–47 (2014).
Roadmap Epigenomics Consortium. et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015).
ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Tentler, J. J. et al. Patient-derived tumour xenografts as models for oncology drug development. Nat. Rev. Clin. Oncol. 9, 338–350 (2012).
Suvà, M. L. et al. Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell 157, 580–594 (2014).
Ricci-Vitiani, L. et al. Tumour vascularization via endothelial differentiation of glioblastoma stem-like cells. Nature 468, 824–828 (2010).
Ricci-Vitiani, L. et al. Mesenchymal differentiation of glioblastoma stem cells. Cell Death Differ. 15, 1491–1498 (2008).
Wang, Z., Martins, A. L. & Danko, C. G. RTFBSDB: an integrated framework for transcription factor binding site analysis. Bioinformatics 32, 3024–3026 (2016).
Bhat, K. P. L. et al. Mesenchymal differentiation mediated by NF-κB promotes radiation resistance in glioblastoma. Cancer Cell 24, 331–346 (2013).
Carro, M. S. et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature 463, 318–325 (2010).
Danko, C. G. et al. Dynamic evolution of regulatory element ensembles in primate CD4+ T cells. Nat. Ecol. Evol. 2, 537–548 (2018).
Luo, X., Chae, M., Krishnakumar, R., Danko, C. G. & Kraus, W. L. Dynamic reorganization of the AC16 cardiomyocyte transcriptome in response to TNFα signaling revealed by integrated genomic analyses. BMC Genomics 15, 155 (2014).
Chuong, E. B., Elde, N. C. & Feschotte, C. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science 351, 1083–1087 (2016).
Wang, M., Zhao, Y. & Zhang, B. Efficient test and visualization of multi-set intersections. Sci. Rep. 5, 16923 (2015).
Stergachis, A. B. et al. Developmental fate and cellular maturity encoded in human regulatory DNA landscapes. Cell 154, 888–903 (2013).
Azofeifa, J. G. et al. Enhancer RNA profiling predicts transcription factor activity. Genome Res. 28, 334–344 (2018).
Canute, G. W. et al. Hydroxyurea accelerates the loss of epidermal growth factor receptor genes amplified as double-minute chromosomes in human glioblastoma multiforme. Neurosurgery 39, 976–983 (1996).
Eller, J. L., Longo, S. L., Hicklin, D. J. & Canute, G. W. Activity of anti-epidermal growth factor receptor monoclonal antibody C225 against glioblastoma multiforme. Neurosurgery 51, 1005–1013 (2002). discussion 1013–1014.
Schmieder, R. & Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 27, 863–864 (2011).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10–12 (2011).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Kuhn, R. M., Haussler, D. & Kent, W. J. The UCSC genome browser and associated tools. Brief. Bioinform. 14, 144–161 (2013).
R Development Core Team R: a language and environment for statistical computing (R Foundation for Statistical Computing, 2010).
Wang, Z., Chu, T., Choate, L. A. & Danko, C. G. Rgtsvm: support vector machines on a GPU in R. Preprint at arXiv [stat.ML] (2017). https://arxiv.org/abs/1706.05544.
Danko, C. G. et al. Signaling pathways differentially affect RNA polymerase II initiation, pausing, and elongation rate in cells. Mol. Cell 50, 212–222 (2013).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome. Biol. 9, R137 (2008).
Hastie, T., Mazumder, R., Lee, J. & Zadeh, R. Matrix completion and low-rank SVD via fast alternating least squares. Preprint at arXiv [stat.ME] (2014). https://arxiv.org/abs/1410.2596.
Jolma, A. et al. DNA-binding specificities of human transcription factors. Cell 152, 327–339 (2013).
Acknowledgements
We thank M. Viapiano and P. Sethupathy for valuable comments on the manuscript and other members of the Danko, Kwak and Lis laboratories for valuable discussions. This research was supported in part by US National Institutes of Health (NIH) grants HG009309 (to C.G.D. and H.K.) and GM25232 (to J.T.L.), and by the Walbridge Foundation for Brain Cancer Research. T.C. thanks the Croucher Foundation for the Croucher Scholarships for Doctoral Study (2013). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or the Walbridge Foundation.
Author information
Authors and Affiliations
Contributions
T.C., Z.W. and C.G.D. analyzed the data. E.J.R., G.B. and H.K. performed molecular experiments. H.K. conceived the chromatin run-on technique, with input from L.J.C. and J.T.L. H.H.S. selected tumors for analysis from the GBM tissue bank. R.J.C. and H.H.S. completed the pathologic analysis. L.S.C. and H.H.S. dissected GBM-15-90 brain tissue. S.L.L. runs the GBM tissue bank and performed the murine xenograft experiments. Data collection and analysis was supervised by C.G.D. The manuscript was written by C.G.D. and T.C., with input from the other authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Suppplementary Information
Supplementary Text and Figures
Supplementary Figures 1–30 and Supplementary Notes 1–7
Supplementary Tables
Supplementary Tables 1–7
Rights and permissions
About this article
Cite this article
Chu, T., Rice, E.J., Booth, G.T. et al. Chromatin run-on and sequencing maps the transcriptional regulatory landscape of glioblastoma multiforme. Nat Genet 50, 1553–1564 (2018). https://doi.org/10.1038/s41588-018-0244-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-018-0244-3
This article is cited by
-
Integrative genome-scale analyses reveal post-transcriptional signatures of early human small intestinal development in a directed differentiation organoid model
BMC Genomics (2023)
-
Spurious intragenic transcription is a feature of mammalian cellular senescence and tissue aging
Nature Aging (2023)
-
PRO-IP-seq tracks molecular modifications of engaged Pol II complexes at nucleotide resolution
Nature Communications (2023)
-
A-MYB and BRDT-dependent RNA Polymerase II pause release orchestrates transcriptional regulation in mammalian meiosis
Nature Communications (2023)
-
RNA polymerase II dynamics shape enhancer–promoter interactions
Nature Genetics (2023)