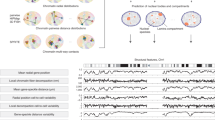
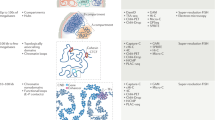
Abstract
Due to recent advances in experimental and theoretical approaches, the dynamic three-dimensional organization (3D) of the nucleus has become a very active area of research in life sciences. We now understand that the linear genome is folded in ways that may modulate how genes are expressed during the basic functioning of cells. Importantly, it is now possible to build 3D models of how the genome folds within the nucleus and changes over time (4D). Because genome folding influences its function, this opens exciting new possibilities to broaden our understanding of the mechanisms that determine cell fate. However, the rapid evolution of methods and the increasing complexity of data can result in ambiguity and reproducibility challenges, which may hamper the progress of this field. Here, we describe such challenges ahead and provide guidelines to think about strategies for shared standardized validation of experimental 4D nucleome data sets and models.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Denker, A. & de Laat, W. The second decade of 3C technologies: detailed insights into nuclear organization. Genes Dev. 30, 1357–1382 (2016).
Van Bortle, K. & Corces, V. G. Nuclear organization and genome function. Annu. Rev. Cell Dev. Biol. 28, 163–187 (2012).
Venter, J. C. et al. The sequence of the human genome. Science 291, 1304–1351 (2001).
Lander, E. S. et al. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001).
Bernstein, B. E. et al. ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Dekker, J. et al. The 4D nucleome project. Nature 549, 219–226 (2017).
Berman, H. M. et al. The Protein Data Bank. Nucleic Acids Res. 28, 235–242 (2000).
Read, R. J. et al. A new generation of crystallographic validation tools for the protein data bank. Structure 19, 1395–1412 (2011).
Westbrook, J., Feng, Z., Burkhardt, K. & Berman, H. M. Validation of protein structures for protein data bank. Methods Enzymol. 374, 370–385 (2003).
Montelione, G. T. et al. Recommendations of the wwPDB NMR validation task force. Structure 21, 1563–1570 (2013).
Sali, A. et al. Outcome of the First wwPDB hybrid/integrative methods task force workshop. Structure 23, 1156–1167 (2015).
Teif, V. B. Nucleosome positioning: resources and tools online. Brief. Bioinform. 17, 745–757 (2016).
Jiang, C. & Pugh, B. F. Nucleosome positioning and gene regulation: advances through genomics. Nat. Rev. Genet. 10, 161–172 (2009).
Buffry, A. D., Mendes, C. C. & McGregor, A. P. The functionality and evolution of eukaryotic transcriptional enhancers. Adv. Genet. 96, 143–206 (2016).
Jin, W. et al. Genome-wide detection of DNase I hypersensitive sites in single cells and FFPE tissue samples. Nature 528, 142–146 (2015).
Buenrostro, J. D. et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523, 486–490 (2015).
Rotem, A. et al. Single-cell ChIP-seq reveals cell subpopulations defined by chromatin state. Nat. Biotechnol. 33, 1165–1172 (2015).
Dion, M. F. et al. Dynamics of replication-independent histone turnover in budding yeast. Science 315, 1405–1408 (2007).
Deal, R. B., Henikoff, J. G. & Henikoff, S. Genome-wide kinetics of nucleosome turnover determined by metabolic labeling of histones. Science 328, 1161–1164 (2010).
Deaton, A. M. et al. Enhancer regions show high histone H3.3 turnover that changes during differentiation. eLife 5, e15316 (2016).
Poorey, K. et al. Measuring chromatin interaction dynamics on the second time scale at single-copy genes. Science 342, 369–372 (2013).
Zentner, G. E., Kasinathan, S., Xin, B., Rohs, R. & Henikoff, S. ChEC-seq kinetics discriminates transcription factor binding sites by DNA sequence and shape in vivo. Nat. Commun. 6, 8733 (2015).
Grimaldi, Y., Ferrari, P. & Strubin, M. Independent RNA polymerase II preinitiation complex dynamics and nucleosome turnover at promoter sites in vivo. Genome Res. 24, 117–124 (2014).
Dekker, J., Rippe, K., Dekker, M. & Kleckner, N. Capturing chromosome conformation. Science 295, 1306–1311 (2002).
Dekker, J., Marti-Renom, M. A. & Mirny, L. A. Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nat. Rev. Genet. 14, 390–403 (2013).
Forcato, M. et al. Comparison of computational methods for Hi-C data analysis. Nat. Methods 14, 679–685 (2017).
Beagrie, R. A. et al. Complex multi-enhancer contacts captured by genome architecture mapping. Nature 543, 519–524 (2017).
Williamson, I. et al. Spatial genome organization: contrasting views from chromosome conformation capture and fluorescence in situ hybridization. Genes Dev. 28, 2778–2791 (2014).
Giorgetti, L. & Heard, E. Closing the loop: 3C versus DNA FISH. Genome Biol. 17, 215 (2016).
Wang, S. et al. Spatial organization of chromatin domains and compartments in single chromosomes. Science 353, 598–602 (2016).
Fudenberg, G. & Imakaev, M. FISH-ing for captured contacts: towards reconciling FISH and 3C. Nat. Methods 14, 673–678 (2017).
Olivares-Chauvet, P. et al. Capturing pairwise and multi-way chromosomal conformations using chromosomal walks. Nature 540, 296–300 (2016).
Quinodoz, S. A. et al. Higher-order inter-chromosomal hubs shape 3D genome organization in the nucleus. Cell 174, 744–757.e24 (2018).
Beliveau, B. J. et al. Single-molecule super-resolution imaging of chromosomes and in situ haplotype visualization using Oligopaint FISH probes. Nat. Commun. 6, 7147 (2015).
Cattoni, D. I. et al. Single-cell absolute contact probability detection reveals chromosomes are organized by multiple low-frequency yet specific interactions. Nat. Commun. 8, 1753 (2017).
Chen, K. H., Boettiger, A. N., Moffitt, J. R., Wang, S. & Zhuang, X. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348, aaa6090 (2015).
Fenrich, K. K. et al. Long-term in vivo imaging of normal and pathological mouse spinal cord with subcellular resolution using implanted glass windows. J. Physiol. (Lond.) 590, 3665–3675 (2012).
Bertrand, E. et al. Localization of ASH1 mRNA particles in living yeast. Mol. Cell 2, 437–445 (1998).
Straight, A. F., Belmont, A. S., Robinett, C. C. & Murray, A. W. GFP tagging of budding yeast chromosomes reveals that protein-protein interactions can mediate sister chromatid cohesion. Curr. Biol. 6, 1599–1608 (1996).
Robinett, C. C. et al. In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J. Cell Biol. 135, 1685–1700 (1996).
Lassadi, I., Kamgoué, A., Goiffon, I., Tanguy-le-Gac, N. & Bystricky, K. Differential chromosome conformations as hallmarks of cellular identity revealed by mathematical polymer modeling. PLOS Comput. Biol. 11, e1004306 (2015).
Tanenbaum, M. E., Gilbert, L. A., Qi, L. S., Weissman, J. S. & Vale, R. D. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell 159, 635–646 (2014).
Saad, H. et al. DNA dynamics during early double-strand break processing revealed by non-intrusive imaging of living cells. PLoS Genet. 10, e1004187 (2014).
Ma, H. et al. Multicolor CRISPR labeling of chromosomal loci in human cells. Proc. Natl. Acad. Sci. USA 112, 3002–3007 (2015).
Chen, B. et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 155, 1479–1491 (2013).
Ma, H., Reyes-Gutierrez, P. & Pederson, T. Visualization of repetitive DNA sequences in human chromosomes with transcription activator-like effectors. Proc. Natl. Acad. Sci. USA 110, 21048–21053 (2013).
Miyanari, Y., Ziegler-Birling, C. & Torres-Padilla, M. E. Live visualization of chromatin dynamics with fluorescent TALEs. Nat. Struct. Mol. Biol. 20, 1321–1324 (2013).
Dubarry, M., Loïodice, I., Chen, C. L., Thermes, C. & Taddei, A. Tight protein-DNA interactions favor gene silencing. Genes Dev. 25, 1365–1370 (2011).
Lenstra, T. L., Coulon, A., Chow, C. C. & Larson, D. R. Single-molecule I=imaging reveals a switch between spurious and functional ncRNA transcription. Mol. Cell 60, 597–610 (2015).
Masui, O. et al. Live-cell chromosome dynamics and outcome of X chromosome pairing events during ES cell differentiation. Cell 145, 447–458 (2011).
Chubb, J. R., Boyle, S., Perry, P. & Bickmore, W. A. Chromatin motion is constrained by association with nuclear compartments in human cells. Curr. Biol. 12, 439–445 (2002).
Hocine, S., Raymond, P., Zenklusen, D., Chao, J. A. & Singer, R. H. Single-molecule analysis of gene expression using two-color RNA labeling in live yeast. Nat. Methods 10, 119–121 (2013).
Chao, J. A., Yoon, Y. J. & Singer, R. H. Imaging translation in single cells using fluorescent microscopy. Cold Spring Harb. Perspect. Biol. 4, a012310 (2012).
Stasevich, T. J. et al. Regulation of RNA polymerase II activation by histone acetylation in single living cells. Nature 516, 272–275 (2014).
Ray-Gallet, D. et al. Dynamics of histone H3 deposition in vivo reveal a nucleosome gap-filling mechanism for H3.3 to maintain chromatin integrity. Mol. Cell 44, 928–941 (2011).
Ricci, M. A., Manzo, C., García-Parajo, M. F., Lakadamyali, M. & Cosma, M. P. Chromatin fibers are formed by heterogeneous groups of nucleosomes in vivo. Cell 160, 1145–1158 (2015).
Matsuda, A. et al. Condensed mitotic chromosome structure at nanometer resolution using PALM and EGFP- histones. PLoS One 5, e12768 (2010).
Abrahamsson, S. et al. Fast multicolor 3D imaging using aberration-corrected multifocus microscopy. Nat. Methods 10, 60–63 (2013).
Abrahamsson, S. et al. MultiFocus polarization microscope (MF-PolScope) for 3D polarization imaging of up to 25 focal planes simultaneously. Opt. Express 23, 7734–7754 (2015).
Hajj, B., El Beheiry, M. & Dahan, M. PSF engineering in multifocus microscopy for increased depth volumetric imaging. Biomed. Opt. Express 7, 726–731 (2016).
Oudjedi, L. et al. Astigmatic multifocus microscopy enables deep 3D super-resolved imaging. Biomed. Opt. Express 7, 2163–2173 (2016).
Ou, H. D. et al. ChromEMT: visualizing 3D chromatin structure and compaction in interphase and mitotic cells. Science 357, eaag0025 (2017).
Wolff, G., Hagen, C., Grünewald, K. & Kaufmann, R. Towards correlative super-resolution fluorescence and electron cryo-microscopy. Biol. Cell 108, 245–258 (2016).
Titze, B. & Genoud, C. Volume scanning electron microscopy for imaging biological ultrastructure. Biol. Cell 108, 307–323 (2016).
Giorgetti, L. et al. Predictive polymer modeling reveals coupled fluctuations in chromosome conformation and transcription. Cell 157, 950–963 (2014).
Gotta, M. et al. The clustering of telomeres and colocalization with Rap1, Sir3, and Sir4 proteins in wild-type Saccharomyces cerevisiae. J. Cell Biol. 134, 1349–1363 (1996).
Pollex, T., Piolot, T. & Heard, E. Live-cell imaging combined with immunofluorescence, RNA, or DNA FISH to study the nuclear dynamics and expression of the X-inactivation center. Methods Mol. Biol. 1042, 13–31 (2013).
Bystricky, K., Laroche, T., van Houwe, G., Blaszczyk, M. & Gasser, S. M. Chromosome looping in yeast: telomere pairing and coordinated movement reflect anchoring efficiency and territorial organization. J. Cell Biol. 168, 375–387 (2005).
Heun, P., Laroche, T., Shimada, K., Furrer, P. & Gasser, S. M. Chromosome dynamics in the yeast interphase nucleus. Science 294, 2181–2186 (2001).
Marti-Renom, M. A. & Mirny, L. A. Bridging the resolution gap in structural modeling of 3D genome organization. PLOS Comput. Biol. 7, e1002125 (2011).
Mirny, L. A. The fractal globule as a model of chromatin architecture in the cell. Chromosome Res. 19, 37–51 (2011).
Barbieri, M. et al. A model of the large-scale organization of chromatin. Biochem. Soc. Trans. 41, 508–512 (2013).
Barbieri, M. et al. Active and poised promoter states drive folding of the extended HoxB locus in mouse embryonic stem cells. Nat. Struct. Mol. Biol. 24, 515–524 (2017).
Fudenberg, G. et al. Formation of chromosomal domains by loop extrusion. Cell Rep. 15, 2038–2049 (2016).
Jost, D., Carrivain, P., Cavalli, G. & Vaillant, C. Modeling epigenome folding: formation and dynamics of topologically associated chromatin domains. Nucleic Acids Res. 42, 9553–9561 (2014).
Emanuel, M., Radja, N. H., Henriksson, A. & Schiessel, H. The physics behind the larger scale organization of DNA in eukaryotes. Phys. Biol. 6, 025008 (2009).
Hahnfeldt, P., Hearst, J. E., Brenner, D. J., Sachs, R. K. & Hlatky, L. R. Polymer models for interphase chromosomes. Proc. Natl. Acad. Sci. USA 90, 7854–7858 (1993).
Münkel, C. & Langowski, J. Chromosome structure predicted by a polymer model. Phys. Rev. E Stat. Phys. Plasmas Fluids Relat. Interdiscip. Topics 57, 5888 (1998).
Di Stefano, M., Rosa, A., Belcastro, V., di Bernardo, D. & Micheletti, C. Colocalization of coregulated genes: a steered molecular dynamics study of human chromosome 19. PLoS Comput. Biol. 9, e1003019 (2013).
Sanborn, A. L. et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc. Natl. Acad. Sci. USA 112, E6456–E6465 (2015).
Chiariello, A. M., Annunziatella, C., Bianco, S., Esposito, A. & Nicodemi, M. Polymer physics of chromosome large-scale 3D organisation. Sci. Rep. 6, 29775 (2016).
Brackley, C. A., Johnson, J., Kelly, S., Cook, P. R. & Marenduzzo, D. Simulated binding of transcription factors to active and inactive regions folds human chromosomes into loops, rosettes and topological domains. Nucleic Acids Res. 44, 3503–3512 (2016).
Gibcus, J. H. et al. A pathway for mitotic chromosome formation. Science 359, eaao6135 (2018).
Stevens, T. J. et al. 3D structures of individual mammalian genomes studied by single-cell Hi-C. Nature 544, 59–64 (2017).
Nagano, T. et al. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature 502, 59–64 (2013).
Flyamer, I. M. et al. Single-nucleus Hi-C reveals unique chromatin reorganization at oocyte-to-zygote transition. Nature 544, 110–114 (2017).
Tan, L., Xing, D., Chang, C.-H., Li, H. & Xie, X. S. Three-dimensional genome structures of single diploid human cells. Science 361, 924–928 (2018).
Serra, F. et al. Restraint-based three-dimensional modeling of genomes and genomic domains. FEBS Lett. 589(20 Pt A), 2987–2995 (2015).
Jhunjhunwala, S. et al. The 3D structure of the immunoglobulin heavy-chain locus: implications for long-range genomic interactions. Cell 133, 265–279 (2008).
Ferraiuolo, M. A. et al. The three-dimensional architecture of Hox cluster silencing. Nucleic Acids Res. 38, 7472–7484 (2010).
Duan, Z. et al. A three-dimensional model of the yeast genome. Nature 465, 363–367 (2010).
Umbarger, M. A. et al. The three-dimensional architecture of a bacterial genome and its alteration by genetic perturbation. Mol. Cell 44, 252–264 (2011).
Baù, D. et al. The three-dimensional folding of the α-globin gene domain reveals formation of chromatin globules. Nat. Struct. Mol. Biol. 18, 107–114 (2011).
Kalhor, R., Tjong, H., Jayathilaka, N., Alber, F. & Chen, L. Genome architectures revealed by tethered chromosome conformation capture and population-based modeling. Nat. Biotechnol. 30, 90–98 (2011).
Trussart, M. et al. Defined chromosome structure in the genome-reduced bacterium Mycoplasma pneumoniae. Nat. Commun. 8, 14665 (2017).
Trussart, M. et al. Assessing the limits of restraint-based 3D modeling of genomes and genomic domains. Nucleic Acids Res. 43, 3465–3477 (2015).
Bianco, S. et al. Polymer physics predicts the effects of structural variants on chromatin architecture. Nat. Genet. 50, 662–667 (2018).
Gerstein, M. B. et al. Architecture of the human regulatory network derived from ENCODE data. Nature 489, 91–100 (2012).
Rao, S. S. et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014).
Heidari, N. et al. Genome-wide map of regulatory interactions in the human genome. Genome Res. 24, 1905–1917 (2014).
Gaspard, N. et al. An intrinsic mechanism of corticogenesis from embryonic stem cells. Nature 455, 351–357 (2008).
Dixon, J. R. et al. Chromatin architecture reorganization during stem cell differentiation. Nature 518, 331–336 (2015).
Bonev, B. et al. Multiscale 3D genome rewiring during mouse neural development. Cell 171, 557–572.e24 (2017).
Stadhouders, R. et al. Transcription factors orchestrate dynamic interplay between genome topology and gene regulation during cell reprogramming. Nat. Genet. 50, 238–249 (2018).
Le Dily, F. et al. Distinct structural transitions of chromatin topological domains correlate with coordinated hormone-induced gene regulation. Genes Dev. 28, 2151–2162 (2014).
Schwarzer, W. et al. Two independent modes of chromatin organization revealed by cohesin removal. Nature 551, 51–56 (2017).
Nora, E. P. et al. Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell 169, 930–944.e22 (2017).
Javierre, B. M. et al. Lineage-specific genome architecture links enhancers and non-coding disease variants to target gene promoters. Cell 167, 1369–1384.e19 (2016).
Hu, J. et al. Direct activation of human and mouse Oct4 genes using engineered TALE and Cas9 transcription factors. Nucleic Acids Res. 42, 4375–4390 (2014).
Qi, L. S. et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173–1183 (2013).
Yan, Q. et al. Multiplex CRISPR/Cas9-based genome engineering enhanced by Drosha-mediated sgRNA-shRNA structure. Sci. Rep. 6, 38970 (2016).
Wijchers, P. J. et al. Cause and consequence of tethering a SubTAD to different nuclear compartments. Mol. Cell 61, 461–473 (2016).
Deng, W. et al. Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell 149, 1233–1244 (2012).
Therizols, P. et al. Chromatin decondensation is sufficient to alter nuclear organization in embryonic stem cells. Science 346, 1238–1242 (2014).
Bantignies, F. et al. Polycomb-dependent regulatory contacts between distant Hox loci in Drosophila. Cell 144, 214–226 (2011).
Deng, W. & Blobel, G. A. Detecting long-range enhancer-promoter interactions by quantitative chromosome conformation capture. Methods Mol. Biol. 1468, 51–62 (2017).
Wilkinson, M. D. et al. The FAIR guiding principles for scientific data management and stewardship. Sci. Data 3, 160018 (2016).
Alber, F., Förster, F., Korkin, D., Topf, M. & Sali, A. Integrating diverse data for structure determination of macromolecular assemblies. Annu. Rev. Biochem. 77, 443–477 (2008).
Goodstadt, M. & Marti-Renom, M. A. Challenges for visualizing three-dimensional data in genomic browsers. FEBS Lett. 591, 2505–2519 (2017).
Acknowledgements
We thank the community of researchers around the EU 4DNucleome Initiative for their continuous support.
Author information
Authors and Affiliations
Contributions
All authors wrote and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Marti-Renom, M.A., Almouzni, G., Bickmore, W.A. et al. Challenges and guidelines toward 4D nucleome data and model standards. Nat Genet 50, 1352–1358 (2018). https://doi.org/10.1038/s41588-018-0236-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-018-0236-3
This article is cited by
-
Gene regulatory network inference in the era of single-cell multi-omics
Nature Reviews Genetics (2023)
-
Understanding 3D genome organization by multidisciplinary methods
Nature Reviews Molecular Cell Biology (2021)
-
The existence of a nonclassical TCA cycle in the nucleus that wires the metabolic-epigenetic circuitry
Signal Transduction and Targeted Therapy (2021)
-
Insulator-based loops mediate the spreading of H3K27me3 over distant micro-domains repressing euchromatin genes
Genome Biology (2020)
-
DeepC: predicting 3D genome folding using megabase-scale transfer learning
Nature Methods (2020)