Abstract
The maize W22 inbred has served as a platform for maize genetics since the mid twentieth century. To streamline maize genome analyses, we have sequenced and de novo assembled a W22 reference genome using short-read sequencing technologies. We show that significant structural heterogeneity exists in comparison to the B73 reference genome at multiple scales, from transposon composition and copy number variation to single-nucleotide polymorphisms. The generation of this reference genome enables accurate placement of thousands of Mutator (Mu) and Dissociation (Ds) transposable element insertions for reverse and forward genetics studies. Annotation of the genome has been achieved using RNA-seq analysis, differential nuclease sensitivity profiling and bisulfite sequencing to map open reading frames, open chromatin sites and DNA methylation profiles, respectively. Collectively, the resources developed here integrate W22 as a community reference genome for functional genomics and provide a foundation for the maize pan-genome.
Similar content being viewed by others
Main
The maize W22 genome has served as a foundation for maize genetics since its early adoption by Brink for studies of paramutation1. The inbred, developed at the Wisconsin Agricultural Experiment Station, lacked the ubiquitous suppressors of anthocyanin pigmentation present in most standard corn belt inbreds but did carry mutations in two regulatory genes controlling anthocyanin production, R1 and C1. Thus, Brink introgressed functional alleles of R1 and C1 to produce a color-converted W22 inbred (PI accession 674445). This inbred was soon adopted by Nelson, Kermicle and Dooner and their students, who led foundational studies to understand the genetics of high-protein maize (O2)2, provide the first example of imprinting3, conduct the first transposon tagging experiments in plants4 and understand the mechanisms of Activator and Dissociation5,6 transposition. Today, the W22 genome is the host to thousands of mobilized, indexed Mutator7,8,9, Activator10,11 and Dissociation12,13 insertions that are used extensively for functional genomics studies. Maize exhibits exceptional levels of structural variation that influences gene content and gene regulation among different inbred lines14,15,16,17. This limits the utility of the B73 reference genome18 or the PH207 genome19 for interpreting data derived for mutant alleles in a W22 genetic background. Detailed knowledge of the W22 genome structure and content is critical to fully exploit reverse genetics resources and biological processes in W22.
W22 genome de novo assembly and validation
The W22 genome was sequenced and assembled using deep sequencing ( > 180×) of Illumina short-read sequences from libraries with a variety of insert sizes using the assembly algorithms developed by NRGene19 (see Methods and Reporting Summary for details). Sequence contigs were developed based on paired-end (PE) short reads and then were put into scaffolds using PE or mate-pair (MP) links. Further ordering and orientation of scaffolds was achieved through alignment to the B73 genome assembly. The initial assembly was improved through utilization of the 10× genomics library platform20 and sequencing using the HiSeq X10 to bridge contigs and correct initial misassemblies (see Methods). This revealed 34 potential chimeric scaffolds that were broken and reassembled to generate a W22v2 genome (Table 1; see Methods). The W22 whole genome is available at NCBI under the accession GCA_001644905.2. The genome and accompanying resources are available at MaizeGDB under the name Zm-W22-REFERENCE-NRGENE-2.0 with the identifier Zm00004b.
Several approaches were used to assess the quality and completeness of the W22 genome sequence at macro and micro scales. To confirm the overall quality of the sequence assembly, we created an independent optical map based on the restriction site BspQI21 and aligned the optical map with an in silico digested version of the genome assembly. The two assemblies strongly supported each other, showing exceptional (>98%) alignment and colinearity. Further confirmation of the assembly was performed using a suite of 4.4 million SNP markers validated against the maize nested association mapping panel22. A genetic assessment of contig and scaffold ordering was conducted by aligning 4.4 million SNP markers to an initial W22 assembly scaffold revealing ~30 inconsistencies (see Methods). This genetic map was used to correct the initial assembly to produce a W22v2 assembly that integrates the genetic and physical maps and provided evidence for robust assembly. The W22v2 genome has a larger number of gaps (as defined by >10Ns) than the B73v4 genome but substantially fewer gaps than the PH207 genome (Supplementary Table 1).
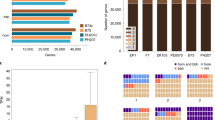
A more granular assessment of the assembly quality and accuracy was made by comparing the W22v2 assembly to a 238-kilobase (kb) interval encompassing the W22-bz1 locus on chromosome 9 that had previously been sequenced using Sanger sequencing technology23. The vast majority (~113 Mb) of this region aligned with > 99% identity in a collinear fashion (Fig. 1). The primary difference between the W22v2 genome assembly and the earlier Sanger BAC assembly is an unfilled gap present in the W22v2 assembly that corresponds to 7,137 base pairs (bp) of a Misfit retrotransposon that is located in the midst of an 85-kb retrotransposon cluster (Fig. 1). There are three other small gaps in this region that occur within repetitive elements but the size of these gaps is similar to the BAC sequence and the order and orientation is the same as the BAC. This analysis provides further evidence for the quality of the W22v2 assembly and suggests that remaining gaps likely occur within repetitive regions.
Benchmarking the W22v2 assembly quality and completeness. A comparison of the W22v2 assembly with a Sanger-based assembly of a 238-kb maize BAC23 (Genbank; EU338354). This region contains five genes (yellow/tan symbols along line) as well as a number of transposons (colored triangles). The W22v2 assembly matches the majority of the sequence at quite high identity (>99%) and the locations of gaps in the W22v2 genome (indicated by red dotted lines).
Annotation of genes and transposons in W22
Genes of the W22v2 genome were annotated using a modified Maker P approach that was developed for B73v418 together with RNA-seq from W22 tissues and full-length cDNA data (see Methods for details). A ‘working gene set’ of 57,181 genes was identified. Removal of putative transposons, low-confidence single-exon genes and potential pseudogenes resulted in a ‘filtered gene set’ of 40,789 genes. The exons of the filtered gene set account for 2.99% and the introns account for another 5.15% of the 2.2 Gb W22v2 assembly. The number of genes and distribution along the chromosomes (Fig. 2) are very similar to previous maize genome assemblies for B734 and PH20719. The quality and completeness of the W22 genome was assessed through a BUSCO analysis24. Over 90% of BUSCO genes are represented by a complete single copy in the W22 genome and only 3% of BUSCO genes are missing in the W22v2 assembly (Supplementary Fig. 1). These rates are equivalent to the B73v4 genome (Supplementary Fig. 1), suggesting similar qualities of annotation completeness and quality in these genomes.
Distribution of features within the W22 genome. The relative density for a variety of features in 1 Mb bins across the maize genome is shown. The purple tracks show the density of genes, LTR transposons and TIR transposons. The relative chromatin accessibility as assessed by MNase (green) and the levels of CG, CHG and CHH methylation (red) are shown. The frequencies of Ds or Mu transposon insertions are also shown (gray).
The availability of a de novo assembly for the W22 genome allows for comparisons of gene copy number, structure and splicing between W22 and B73. Previous work has found evidence for frequent copy number variation and presence/absence variation among maize inbreds17,19,25. The B73 and W22 genomes were compared to each other and to the Sorghum BTx623 genome26 to document the presence or absence of syntenic orthologs among these three genomes (Supplementary Table 2). Over 60% of the genes are present at syntenic locations in both maize lines and Sorghum. Another 15–20% of genes are found in both B73 and W22 but are absent from the BTx623 assembly (Supplementary Table 2). The remaining genes are present in one maize line and absent in the other. The majority of these genes are present only in B73 (6,440) or W22 (8,372) and not found in the other maize inbred or in Sorghum. There are relatively fewer examples of genes that are present in Sorghum and only one maize inbred (B73, 881; W22, 1,412). The presence of locally duplicated genes was evaluated in both the W22 and B73 genome, for a range of intervening genes from zero (tandem duplicates) to twenty (Supplementary Tables 3 and 4). A subset (678) of the examples of tandem duplications are observed in both B73 and W22. However, there are many other cases of tandem duplication that are observed only in B73 (1,394) or in W22 (1,261). The tandem duplicates were classified according to their relative arrangement (Supplementary Table 5). Tandem duplicates in the same orientation (head to tail) are more common than genes in a convergent (tail-to-tail) or divergent (head-to-head) arrangement (Supplementary Table 5).
The presence of a tandem duplication in one genome and not in the other could reflect mechanisms to rapidly expand or contract gene copy variants and may serve as a foundation for functional diversification; for example, the terpene synthase family has variable tandem duplications that may affect function (see Supplementary Note for details).
RNA-seq data generated from several W22 tissues were used to document examples of alternative splicing in W22 and compare these alternative splicing events to those observed in B7327. We identified 13,591 W22 genes with alternative splicing, representing 49.4% of expressed multi-exon genes. The types, and frequencies, of alternative splicing events noted in W22 (Supplementary Table 6) are similar to those noted for B7327, albeit at lower numbers, which may be due to reduced levels of RNA-seq data for W22 relative to B73. There are numerous locus-specific differences in splicing observed between B73 and W22 and these are found distributed throughout the genome (Supplementary Fig. 2). One major advantage of sequencing and annotating additional genotypes is the improved ability to analyze high-throughput sequencing data sets. To test the benefit of using a genotype-matched reference, W22 leaf RNA sequencing data (SRA: SRR1986376) was analyzed in parallel using both the W22 and the B73v4 genome references. Mapping RNA-seq reads from W22 seedling leaf tissue to the W22 genome resulted in a mapping rate of 95.7% while only 91.1% of these reads could be aligned to the B73v4 genome (Supplementary Fig. 3a). A comparison of the expression level for orthologous genes in B73 and W22 reveals that the expression level estimates vary for a number of genes (Supplementary Fig. 3b). In many cases, these changes in expression were due to altered annotation of the transcripts in W22 relative to B73 (Supplementary Fig. 3c).
The maize genome is largely composed of transposable elements (TEs)28. A structural annotation of transposons in the W22v2 genome identified over 177,000 transposons that were classified into 26,833 families (see Methods). Long terminal repeat (LTR) retrotransposons are the most prevalent type of TEs in the W22 genome with 23,144 families accounting for 64% of the genome. The 10 most abundant LTR families have 1,400–16,395 members and account for 36% of the genome. Terminal inverted repeat (TIR) DNA transposons were classified into 5 major types and account for 0.46% of the genome. There are also 23,895 helitron elements that account for 4.6% of the genome. While many of the TEs are present in intergenic space, there are numerous examples of transposons being annotated within the introns of genes. Nearly 9% of the FGS genes contain an annotated transposon, including 1,626 genes containing a TIR element and 1,864 genes containing an LTR element. A comparison of the TEs present in the B73 and W22 genome revealed high levels of variation in TE presence and copy number for both TIR and LTR families. There are 937 families of TIR elements that are found in both B73 and W22. Many other TIR families are found only in B73 (107 families) or W22 (62 families) after excluding B73 families that are present only as MITEs. These B73-specific and W22-specific TIR elements are enriched for CACTA (DTC) families (Supplementary Fig. 4a). There are 12,740 families of LTR elements annotated in both B73 and W22 and many other families specific to W22 (10,531) or B73 (11,032). The number of elements in each family is generally similar in the two genomes but there are many examples of strong copy number variation (Supplementary Fig. 4b–d). However, there are 23 LTR families and 30 TIR families with differences in copy number >100 between the two genomes including 27 with higher copy number in W22 and 26 with higher copy number in B73. The analysis of age of LTR elements (based on sequence identity of the two LTRs) in families that are inbred-specific reveals that small inbred-specific families tend to be relatively old and thus may be being purged from the genome while larger inbred specific families tend to be enriched for recent insertion events (Supplementary Fig. 4e). Even in TIR and LTR families with similar copy number we see variation for specific elements. For example, among the 100 TIR families with a single copy in each genome there are 74 examples in which the transposon is inserted into different chromosomes in the two inbred lines. A detailed analysis of native Mutator insertions in the B73 and W22 genomes (see Supplementary Note for details) revealed similar numbers in both genomes (258 in W22 and 259 in B73). However, fewer than half (125) of these insertions are shared in both lines. There are over 100 unique Mutator insertions in each line and 63% of these are located within 500 bp of an annotated gene. Among the 7,463 LTR families with a single copy in both B73 and W22 there are 863 examples in which that single element is located on different chromosomes in the two genomes. These examples of polymorphic TIR and LTR elements highlight the exceptional diversity of TE insertion events among maize genotypes.
Mapping of transposon insertion sites using the W22 genome
Significant efforts have been dedicated to the development of stocks with novel transposon insertions in the W22 genetic background. These include several approaches that have mobilized Ds elements13 and the development a large number of Mu insertion stocks7,8,9. A large number of these novel insertions have been sequence indexed to document information on the sequence flanking insertion sites. To date, the insertion sites have often been determined through mapping of flanking sequences to the B73 genome. However, a large number of insertions could not be accurately placed due to insufficient sequence identity with B73. To precisely position the 2,029 Ds and 68,866 Mu insertions that have been generated, sequences adjacent to the transposon were mapped against the W22v2 genome. This provided novel insertion sites for 12%–15% of these insertions (Fig. 3). For the 13,444 UniformMu stocks, the mapping of flanking sites to the W22 genome provided novel insertion site information for over 10,000 insertions and increased the properly placed insertions from 4.35 per line to 5.1 per line. Furthermore, the genomic distributions of Ds and Mu insertions are distinct from one another (Fig. 2), indicating complementarity of these resources for functional genomics studies. The improved mapping resolution of these insertions coupled with a better resolution of gene content and position afforded by W22v2 assembly will greatly increase the utility of the Ds and Mu reverse genetics resources. In particular, the Ds resource is designed for remobilization of elements to flanking regions of the genome, so accurate placement of insertions to gene models and local gene content is imperative for efficient mutagenesis. Similarly, better gene models will enable a more accurate prediction of phenotypic consequences of transposon insertions, particularly those that map to the 5ʹ and 3ʹ ends of a gene and thus may induce subtle mutant phenotypes.
Improved ability to document the location of Ds and Mu insertions using the W22 genome. The Ds (n = 2,029) and Mu (n = 68,866) insertion flanking sequences were mapped to the W22 genome. The proportion of insertion sites that mapped to the same location (black), different locations (blue) or only to W22 (red) was determined.
Ds and Mu probe different compartments of the genome
Ds and Mu represent two different transposon superfamilies (DTA and DTM). Previous studies have suggested differences in terms of the frequency of linked transposition and insertion locations relative to genes13,29,30. Several properties of chromatin in the W22 genome were analyzed to better understand the factors that influence the insertion sites for Ds and Mu. The context-specific DNA methylation patterns throughout the W22 genome were documented using whole-genome bisulfite sequencing (WGBS) and chromatin accessibility was assessed using MNase treatments (Figs. 2 and 4a–c). The levels and genomic distribution of DNA methylation in W22 are similar to patterns observed in other maize inbred lines31,32,33. Open chromatin and CHH methylation are substantially enriched in promoter regions and following the transcription termination sites of annotated genes (Fig. 4a). In contrast, CHG and CG methylation are relatively depleted in these regions (Fig. 4a). CG and CHG methylation are enriched within TIR and LTR transposons (Fig. 4b,c). CHH methylation is enriched at TIR elements but shows minimal enrichment at LTR elements. Chromatin accessibility does not vary substantially within or near transposons in the W22 genome (Fig. 4b,c).
Patterns of chromatin over maize genes and transposons. a–c, The levels of CG methylation (black), CHG methylation (blue) or CHH methylation (red) over the 1 kb flanking regions and maize genes (a), TIR transposons (b) or LTR transposons (c). In addition, the enrichment for open chromatin (green) based on MNase digests is shown for the same regions. d–f, Chromatin profiles for sites targeted by Ds (solid lines) or Mu (dashed lines). The relative levels of CG methylation (d) and CHG methylation (e) or open chromatin (f) are shown for the flanking regions (up to 10 kb) near Ds or Mu insertion sites.
The locations for novel transposon insertions can be quite sensitive to chromatin structure34. The profiles of open chromatin and DNA methylation were assessed for the regions flanking potential Ds and Mu insertion sites (Fig. 4d–f and Supplementary Fig. 5). There are differences in the accessibility of chromatin at Mu and Ds insertion sites. The Mu insertion sites are marked by an increase in chromatin accessibility while Ds insertion sites do not vary substantially compared to flanking regions. Both Mu and Ds prefer to insert in regions with very low CG and CHG methylation. A metaprofile from bisulfite sequencing reveals that Mu insertion sites are often very near regions with CG or CHG methylation as both of these modifications are present on either side of the insertion site. In contrast, the increase in CG or CHG methylation near Ds insertion sites tends to occur further away from the insertion site. CHH methylation tends to be enriched at regions immediately adjacent to Mu insertion sites but does not show strong patterns near Ds insertion sites (Supplementary Fig. 5). These patterns could explain the preference for Mu to insert in the 5ʹ and 3ʹ UTR regions of genes while Ds is more likely to insert within coding regions.
The W22 genome contains a number of native TIR transposon families. The patterns of chromatin within these TIR elements include elevated DNA methylation and relatively closed chromatin (Fig. 4b). The chromatin at the regions flanking these TIR elements likely reflects endogenous genomic information that influences chromatin state as well as the influences of the TE on neighboring chromatin. Given the differences in chromatin profiles for the insertion sites of Ds or Mu, we were interested in comparing the chromatin at these regions with the chromatin flanking pre-existing TIR insertions. We collected chromatin profiles for 2 kb of flanking regions for all TIR families with >50 elements and clustered these profiles together with the profile of chromatin flanking potential Ds and Mu insertion sites (Fig. 5). There are several quite distinct patterns of chromatin flanking TIR elements. Some families show very high methylation for flanking regions while other families are more likely to have low levels of CG and CHG methylation. The Mu and Ds potential insertion sites are outliers for the levels of CG and CHG methylation compared to the regions flanking pre-existing element insertion sites. The analysis of the methylation levels for the first 100 bp at the edge of the TE reveals that both Mu and Ds insertion sites have much lower levels of CG and CHG methylation than observed for pre-existing TIR insertion sites (Fig. 5b,c). In contrast, the levels of CHH methylation near Mu and Ds potential insertion sites are very similar to the levels of CHH methylation at pre-existing TIR insertion sites (Fig. 5d).
Chromatin profiles for regions flanking pre-existing TIR elements or novel Ds or Mu insertion sites. a, For 191 TIR families with at least 20 elements present in the W22 genome, we determined the relative level of CG and CHG methylation in 100-bp windows for the 1-kb flanking regions. These levels of methylation were used to cluster the families and a heatmap allows visualization of the patterns of DNA methylation for flanking regions. b,c, The distributions of the average level of methylation for CG (b) or CHG (c) and CHH (d) contexts in the 100 bp surrounding each TE family are shown as density plots. The levels of methylation at sites where Mu (red arrows) or Ds (black arrows) elements can insert are shown as arrows.
Since the turn of the twentieth century, maize has provided a model system for genetic research35. The development of a color-converted W22 inbred enabled high-resolution mapping of numerous traits influencing anthocyanin and flavonoid accumulation and led to the adoption of W22 as the workhorse of maize genetics1,2,3,4. In recent years the genomics data sets generated from studies of transposon insertion alleles have been analyzed in the context of the B73 reference genome. Numerous studies have documented the widespread genetic variation among inbred genotypes of maize including SNPs, copy number variation and indels15,19,36,37. The availability of a high-quality assembly and annotation for W22 will increase the resolution of studies of mutant alleles in this genetic background and recent improvements in maize transformation are likely to enable higher efficiencies of W22 transformation38. Thus, W22 is well positioned to serve as the maize reference inbred for functional genomics studies.
The annotation of TEs present in the W22 and B73 genome will provide new opportunities to study the sources, and effects, of TE polymorphisms among maize lines. While there are many shared transposons in B73 and W22, there are numerous examples of severe copy number and presence/absence variation of certain families. There are also many examples of polymorphic TE insertions within or near genes that could influence regulation of these genes. Locus-specific studies have provided evidence for the importance of polymorphic TE insertions that influence the regulation of nearby genes, resulting in quantitative trait loci affecting important agronomic traits39,40,41. The ability to have a high structural resolution of both genomes will serve as a resource to enable detailed studies of how transposons influence gene expression among diverse maize genotypes. The knowledge of complex haplotype variation can also be used to inform pan-genome-based approaches that incorporate rearrangements and insertion/deletions from large-scale resequencing studies to augment SNP-based GWAS studies.
The availability of epigenome profiles for DNA methylation and chromatin accessibility provides an opportunity to study chromatin profiles for thousands of Mu and Ds insertion sites. The differences in these profiles suggest that Mu and Ds utilize distinct targeting mechanisms and occupy different niches of the W22 genome. Mu elements tend to insert in small windows of accessible chromatin that are near highly methylated regions. Previous work has noted the propensity for Mu elements to insert near gene promoters and these regions likely contain the chromatin profiles noted for Mu elements29,30. In contrast, Ds frequently inserts into regions with low CG and CHG methylation that are not enriched for accessible chromatin. This is compatible with the concept of preferential insertion of Ds elements within the coding regions of maize genes13. Interestingly, chromatin profiles for random sites selected within gene promoters or coding regions do not match the profiles observed for Mu or Ds sites. This suggests that Mu and Ds have specific targeting beyond simple selection for promoters or coding regions. Furthermore, the chromatin profiles at loci targeted by active Mu or Ds elements are distinct from the chromatin profiles at sites that flank inactive pre-existing insertion sites. This likely reflects the fact that many of these pre-existing element insertion sites are targeted for silencing and the chromatin properties of the elements themselves can spread into the flanking regions. It is possible that many of the pre-existing TIR families might insert into regions with chromatin profiles similar to Mu or Ds if they were active. As additional high-quality maize genome assemblies become available, comparative analysis of the maize pan-genome is likely to provide further insight into how transposons have shaped the maize genome through the course of domestication.
Methods
W22 genome sequencing and assembly
DNA sequencing
Genomic DNA was isolated from 10-day-old W22 seedling tissues and nuclear DNA was purified and fractionated from organellar DNA on a CsCl gradient42. A genomic library of 800-bp DNA fragment sizes was prepared using the TruSeq DNA Sample Preparation Kit version 2 according to the manufacturer’s protocol (Illumina). A second shotgun library was made using the same kit from DNA template fragments size selected of ~450 bp with no PCR amplification (PCR-free). This fragment size was designed to produce a sequencing overlap of the fragments to be sequenced on the HiSeq2500 v2 Rapid mode as PE sequencing 250 nucleotides per end, thus creating an opportunity to produce ‘stitched’ reads of approximately 250 bp to 480 bp in length. Multiple MP libraries were prepared with the objective to increase sequence diversity and genome coverage including three size classes (2–4 kb, 5–7 kb and 8–10 kb) using the Illumina Nextera Mate-Pair Sample Preparation Kit (Illumina). The 800-bp shotgun library and the MP libraries were sequenced on an Illumina HiSeq2500 as PE with 160-bp read lengths for each of the fragments using the SBS sequencing kit version 4. Each library was sequenced to a coverage of ~65 Gb (~30× the maize genome size). The 450-bp PCR-free shotgun library was sequenced on an Illumina HiSeq2500 using Rapid mode with v2 chemistry as PE with 250-bp read lengths. This library was sequenced to a coverage of ~130 Gb (~60× the maize genome size). In total, ~180× of sequencing data was produced for this project. All sequencing was conducted at the Roy J. Carver Biotechnology Center (Urbana, IL) at the University of Illinois.
Genome assembly
The W22 genome was assembled using DenovoMAGIC 2 (NRGene). Several previous studies have described this assembly approach19,43 and we provide a summary of the implementation of this approach for W22. Read pre-processing included removal of PCR duplicates, Illumina adaptor AGATCGGAAGAGC and Nextera linkers (for MP libraries). For the 2 × 250 450-bp PE libraries, overlapping reads were merged with a minimal required overlap of 10 bp to create stitched reads. Error correction of pre-processed reads was performed by scanning to detect reads with putative sequencing errors (containing a sub-sequence that does not reappear several times in other reads) and those reads were filtered out. The first step of de novo assembly consisted of building a de Bruijn graph (k-mer = 239 bp) of contigs from the ‘stitched’ reads. The large k-mer length significantly reduced the complexity of the de Bruijn graphs, which is essential for high-quality assembly of very complex genomes. No bubble merge and no repeat masking filtering were used.
To scaffold the contigs of the de Bruijn graph, non-repetitive contigs within the graph were identified and assembled into scaffolds based on mapping information of the ‘stitched’ reads. Scaffolding was completed using a directed graph containing scaffolds longer than 200 bp as nodes, and edges were based on the PE and MP links as vertices. Erroneous connections were filtered out to generate unconnected sub-graphs that were ordered into scaffolds. PE reads were used to find reliable paths in the graph for additional repeat resolving. This was accomplished through searching the de Bruijn graph for a unique path connecting pairs of reads mapping to two different scaffolds. The scaffolds were then furthered ordered and linked using the MP libraries, estimating gaps between the contigs according to the distance of MP links. Linking scaffolds with MP reads required confirmation of at least three filtered MPs or at least one filtered MP with supporting confirmation from two or more filter-failed MPs where the Nextera adaptor was not found. Scaffolds shorter than 200 bp were masked and links between non-repetitive contigs mapping to the same scaffolds were united, generating a directed scaffold graph. Further ordering of scaffolds was achieved through alignment of the scaffolds to the B73 reference genome (RefGen_v2) and selection of the most probable genomic location. We did not use the B73 reference to create the W22 scaffolds. The scaffolds were then placed into the pseudomolecules to maximize linear synteny between the assembled and B73 genomes. To improve the accuracy of our scaffolds, we used an independent method to identify and correct misassemblies. We sequenced one HiSeq X10 lane (total of 120G of 150 × 2 bp raw sequencing data) with one long-range barcoded DNA library (Chromium system by 10x genomics). Reads were mapped to the assembled scaffolds and clusters of reads with the same barcode mapped to adjacent contigs in the scaffolds were identified. Overall, we detected 22 million clusters and the estimated length of >2 million of those molecules is above 50 kb (the estimated average length of all molecules is 15 kb with s.d. = 23 kb). Next we scanned along each scaffold with a 20-kb-length window and counted: the number of distinct clusters that cover the left edge of the window; the number of distinct clusters that cover the right edge of the window; and the number of distinct clusters that cover the entire window indicating a support for this 20 kb connection by several long molecules (identified by having distinct barcodes). On average, 255 long molecules (s.d. = 74) cover each edge of the window and 124 long molecules support an entire window (s.d. = 31). There are 34 potential chimeric scaffolds that were detected as windows with a statistically significant (using a hyper-geometric statistical test with a Bonferroni correction, P value 2.5 × 10−7) lower number of supporting overlapping long molecules. Twenty-three of these potential chimeric scaffolds were also detected in the genetic marker-based scaffold validation stage (see below). These chimeric scaffolds were broken and reassembled in W22v2 using the X10 information.
Bionano mapping and assembly validation
High-molecular-weight genomic DNA was isolated from 0.5 g of W22 leaf tissue using the IrysPrep Plant Tissue DNA Isolation Kit (RE-014-05). The DNA was labeled at Nt.BspQI sites using the IrysPrep NRLS labeling kit (RE-012-10). Molecules were collected using BioNano IrysChips at 100× coverage with an average molecule length of 220 kb. The data were then de novo assembled using ‘optArgument_human’. The resulting W22 assembly contained 1,872 BioNano genome maps (equivalent to contigs) that span a total of 2,171 Mb with an N50 of 1.646 Mb. After assembly, the W22v2 genome maps were aligned to an in silico BspQI-digested cmap of the NRGene sequence assembly. The final alignment parameter ‘Total Unique Aligned Len/Ref Len’ was 0.983, indicating that 98.3% of the BioNano maps aligned uniquely with the NRGene assembly. As a means to directly compare the quality of the W22 assembly with the quality of the B73 assembly18, we created a hybrid scaffold that incorporated both the sequence and Bionano data. The W22 hybrid assembly contained 97.4% of the Bionano contigs and 99.4% of the sequence assembly. These values are slightly better than equivalent values for B73 (95.1% and 98.4%, respectively), indicating that the ordering and orientation of contigs in the NRGene assembly are on par with the PacBio-based B73 reference assembly.
Scaffold validation
The 4.4 million high-resolution genetic anchors, which were developed from >14,000 maize inbred lines22, were used to validate the scaffolds of the W22 assembly. The sequence anchors were aligned to an initial assembly of W22 using Bowtie2. The scaffolds with at least 20 mapped anchors were examined for the consistency of sequence assembly and genetic position. A total of 409 scaffolds with an overall length of 2.17 Gb were evaluated. We found that 23 scaffolds were assembly artifacts, which were then corrected in the W22v2 assembly.
W22 genome annotation
W22 gene annotation
Annotation of protein-coding genes was performed using MAKER-P pipeline software44, with parameters and evidence similar to those recently used to annotate B7318,45. Repeat masking (‘RepeatMasker Home Page’ 2017) was performed using exemplar transposon sequences28 available online at the maize TE database (‘Maize Database’ 2017). We excluded helitron and MULE elements to avoid false-positive masking from captured exon sequences in such elements. Gene expression evidence included PacBio Iso-seq long reads sequenced from cDNA libraries of six tissues in B73 (n = 111,151)46. In addition, we included the following transcriptome assemblies, each processed to exclude short transcripts (<300 bp) and redundancies based on application of CD-HIT47: a pooled set of 94 transcriptome assemblies constructed from publicly available RNA-seq reads (n = 508,233)45; a transcriptome assembly of B73 seedlings (n = 112,963)48; a transcriptome assembly of W22 tissues (n = 589,743). Cross-species evidence was supplied in the form of the following annotated protein files downloaded from Gramene release 46(Gramene FTP)49: Arabidopsis_thaliana.TAIR10.27.pep.all.fa;Brachypodium_distachyon.v1.0.27.pep.all.fa; Oryza_sativa.IRGSP-1.0.27.pep.all.fa; Setaria_italica.JGIv2.0.27.pep.all.fa; and Sorghum_bicolor.Sorbi1.27.pep.all.fa. Alignment and downstream processing of sequence evidence to the repeat-masked W22 reference was performed within the MAKER-P pipeline using default parameters. For gene model prediction, the pipeline incorporated AUGUSTUS50 applied with the ‘maize5’ model and FGENESH51 applied with the ‘monocot’ model. Stable gene identifiers were assigned using the format Zm00004bXXXXXX (where the Xs represent a random 6-digit number), as specified under ‘A Standard For Maize Genetics Nomenclature’ available at MaizeGDB (‘Maize Genetics Nomenclature’ 2017). For further details on the characterization and screening of gene models, see the Supplementary Note.
TE annotation
To identify structurally intact copies of TEs in the genome assembly, we followed a similar approach to that used for B73v418 to characterize features such as target site duplications and terminal motifs. To encourage consistency of nomenclature, we applied the 80-80-80 bioinformatic cutoffs52, to match W22 TE copies to existing families in B73 based on 80% identity across 80% of the length of the TE or its functional sequence, of at least 80 base pairs. For TEs that could not be assigned to a family present in B73, we generated new family names. Each TE is given a unique identifier in the form RLG00001Zm00004b00001, using the Zm00004b identifier for W22 assigned by the maize community nomenclature committee for gene names. This reflects the TE superfamily (RLG, retrotransposon, LTR, Gypsy), a five digit family number (00001), the W22 identifier (Zm00004b) and a unique identifier for this TE copy (00001). For LTR elements, we apply an iterative search of LTRharvest53, removing previously identified copies from the genome, and rerunning LTRharvest with the reduced genome. For TIR elements, we utilize TARGeT54, using TE models from the maize TE consortium (MTEC), as well as novel structural families identified using detectMITE55. For helitrons, we use the terminal 30 bp at the 3ʹ end to assign to B73 families, and generate new families as needed. For SINE elements, we use SINEfinder56, and for LINE elements, we use TARGeT on MTEC TE models. The TE detection pipeline and associated files can be found at github.com/mcstitzer/w22_te_annotation.
Characterization of variation in gene content and arrangement
To compare syntenic ortholog retention between W22 and B73, we queried the unmasked W22 genome against the unmasked B73 version 4 from Ensembl release 36 and the outgroup Sorghum bicolor v3.126 using CoGe’s SynFind (lastz, 50-gene window, minimum of 5 syntelogs per window), which compares synteny across multiple genomes at once, and then reciprocally compared B73 against W22 and Sorghum.
Determining locally duplicated genes
Locally duplicated genes were determined using a method similar to that used to determine the number of shared genes between W22 and B73. The CDS sequences from W22 AGPv2 and B73 AGPv4 genomes were each searched against themselves using COGE’s SynFind tool57, and the resulting unfiltered LAST search results were downloaded. In each search, LAST hits were cleaned by: removing hits with an e-value of 0.001 or more; condensing hits from multiple transcripts between gene models, to a single hit between gene models (that is, a hit between two gene models was retained if at least one transcript model from each gene model had a valid hit to each other); and removing hits between gene models that are not on the same chromosome. The remaining non-self LAST hits were treated as mappings between paralogs within each inbred genome and were used to determine locally duplicated genes. Distances between genes in the remaining LAST hits were determined by calculating their order and orientation on the chromosome and determining the number of intervening genes. Any gene pair that had less than a maximum of 20 intervening genes was classified as locally duplicated58.
Shared and unique tandem duplicates in B73 and W22 were determined by using the master list of ortholog mappings using SynFind. Reciprocal SynFind searches were used where W22 was searched against B73 and vice versa. The reciprocal B73 to W22 gene model mappings that were found in both searches were filtered as syntenic orthologs between the two genomes. Tandem duplications were a subset of the locally duplicated genes that had no intervening genes between them.
Analysis of gene expression
RNA-seq alignments
RNA-seq data for W22 leaf (SRA: SRR1986376) were aligned to both the W22 and B73v4 reference genomes. Reads were trimmed using Cutadapt version 1.8.1 and mapped using tophat2 version 2.0.13 to both genomes. Stranded reads were assigned to gene models for each reference using HTSeq version 0.5.3. The counts per gene were normalized to reads per million mapped reads. To compare gene expression values for mappings to the two references, homologus genes were determined from predicted protein sequences for the first isoform of each gene using OrthoFinder version 1.1.8. Only genes with a 1-to-1 match between W22 and B73v4 were retained for comparison.
Alternative splicing analysis
W22 RNA-seq reads were trimmed using Cutadapt 1.7.1 to remove adapters and low quality sequences trimmed using Trimmomatic 0.32. Processed reads were then aligned to the genome using STAR 2.5.2 and two pass alignment steps. Sixty-seven RNA-seq sample were aligned to STAR individuals, and splice junction from 67 samples were combined together and only splice junction > = 3 reads in at least one sample kept. These remaining splice junctions were fed into STAR as guidance and the second alignment was run. Duplicate reads were detected and removed using Picard 2.10.3 MarkDuplicates. Two approaches, Stringtie 1.3.3 and Cufflinks 2.2.1, were used for the genome guided assembly; in addition, we also used de novo transcripts from Trinity. The transcripts built from Cufflinks and Stringtie were filtered using the same approach previously27,59. To compare alternative splicing in W22 and B73, we took the transcripts from the B73v4 annotation that includes alternative splicing isoforms, and mapped back to the W22v2 genomes and called the alternative splicing loci in B73 relative to the W22v2 genome.
Mapping of Mu and Ds insertion sites
Germinal Mu insertions from 13,444 UniformMu lines7,9 were mapped in the B73v4 and W22v2 genomes using the Illumina-based MuSeq protocol as described in an earlier study8. Insertion sites that were assigned unique locations in the W22 genome were analyzed to determine the presence and locations of corresponding sites in the B73v4 genome. Correspondence of insertion sites in W22 and B73 was based on shared MuSeq reads. Insertion sites in W22 identified by MuSeq reads that failed to align to B73 were scored as W22-specific. Flanking Ds (fDs) sequences, which derive from sequenced clones, were placed on each genome assembly using the processing pipeline and placement quality criteria described previously13, with minor modifications. For each genome, the pipeline was first applied to the unmasked genome and unmasked fDs sequences, and then the remaining, unplaced fDs sequences were repeat masked and run through the pipeline again with the unmasked genome. Multiple fDs from the same insertion event were collapsed into a single placement when at least one fDs clone included a sequence directly adjacent to the Ds insertion site. Repeat masking used Repeatmasker as described previously13.
DNA methylation and chromatin accessibility
Whole-genome bisulfite sequencing and analysis
Genomic DNA (1 µg) was sheared to a size of 200–300 bp. These DNA fragments were then used to construct a whole-genome bisulfite sequencing library using the KAPA library preparation kit (KK8232). Briefly, the DNA fragments were subjected to end repair, A-tailing, adapter ligation and dual-SPRI size selection following the manufacturer’s instructions. The resulting library, which has a size between 250 bp and 450 bp, was treated with bisulfite sodium so that unmethylated cytosine could be converted to uracil using the Zymo EZ DNA methylation lightning kit (D5031). The KAPA HiFi HotStart Uracil + (KK2801) was used in the PCR reaction with the following program: 95 °C for 2 min, 8 cycles of 98 °C for 30 s, 60 °C for 30 s, 72 °C for 4 min and a final extension step at 72°C for 10 min. Finally, the PCR-enriched library was cleaned up using SPRI beads. The library was sequenced using Illumina HiSeq2000 with the PE mode and 100 cycles and the reads are available at SRX3136383. Trim_glore was used to trim adapter sequences and read quality was assessed with the default parameters and PE reads mode. Reads that passed quality control were aligned to the W22 assembly using Bismark, allowing at most 1 mismatch in the 25 bp seed sequence (-N 1 -L 25). Duplicate reads were detected and removed using Picard tools. Reads that are uniquely mapped and that are properly paired were used to extract methylation status at each individual cytosine using bismark_methylation_extractor. For PCR inserts that have a size of less than 200 bp (so that the two sequencing reads might overlap), only the first read was used to calculate the methylation level. The file with methylation information for individual cytosines was used to create 100-bp non-overlapping sliding windows across the W22 chromosomes for each of the three sequence contexts, CG, CHG and CHH (H = A, C or T). Within each 100-bp window, the total number of methylated and unmethylated reads for every cytosine was summed for each sequence context, and methylation levels were derived using the following formula: (no. methylated_reads/(no. methylated_reads + no. unmethylated_reads)).
MNase assays and analysis
Seeds were sown into Fafard 3 soil mix in two 11” × 21” × 2.5” plastic trays. Nine days after planting, above-ground tissue was cut at the soil line and flash frozen in two batches (one per tray) and stored at −80 °C until grinding. The two batches of frozen seedlings were ground under liquid nitrogen with a mortar and pestle and crosslinked by stirring for 10 min in 100 ml ice-cold fixation buffer (15 mM Pipes-NaOH at pH 6.8, 0.32 M sorbitol, 80 mM KCl, 20 mM NaCl, 0.5 mM EGTA, 2 mM EDTA, 1 mM dithiothreitol, 0.15 mM spermine and 0.5 mM spermidine) containing 1% formaldehyde. Fixation was stopped by adding glycine to 125 mM. Nuclei were isolated by adding Triton X-100 to 1% final by the addition of 0.1 vol of a 10% (vol/vol) Triton X-100 stock, followed by stirring for 10 min. The suspension was filtered through one layer of Miracloth (Calbiochem) and placed in 50 ml centrifuge tubes. In these centrifuge tubes, 35 ml nuclear suspensions were underlaid with a 15 ml Percoll cushion composed of 50% (vol/vol) Percoll (GE) in BFA. Nuclei suspensions were centrifuged at 3,000g for 15 min at 4 °C. The nuclei at the Percoll interface were transferred to a 50 ml tube and diluted twofold with MNase digestion buffer (50 mM Tris-HCl at pH 7.5, 320 mM sucrose, 4 mM MgCl2 and 1 mM CaCl2). Nuclei suspensions were centrifuged at 2,000g for 10 min at 4 °C, and nuclei pellets were resuspended in 2.5 ml MNase digestion buffer. Nuclei were aliquoted into 500 ml aliquots, flash-frozen in liquid nitrogen and stored at −80 °C. Nuclei were thawed at room temperature and digested by adding MNase to 10 U ml−1 (light) or 100 U ml−1 (heavy), and incubated at room temperature for 5 min. Digestions were stopped with 10 mM EGTA. Nuclei were de-crosslinked by incubation overnight at 65 °C in the presence of 1% SDS and 100 μg ml−1 proteinase K. DNA was extracted by phenol–chloroform extraction followed by ethanol precipitation. Digested DNA was resuspended in 40 μg ml−1 RNAse A and size-selected with a two-sided Ampure XP bead purification step (0.5× beads:DNA followed by 2.5× beads:DNA). Size-selected DNA was used to prepare sequencing libraries using the NEBNext Ultra DNA Library Prep Kit for Illumina (NEB), using the manufacturer's instructions. Indexed libraries were pooled and sequenced on four Illumina HiSeq 2500 lanes with PE 50-cycle sequencing. Data are available in NCBI SRA (accession SRP118121).
Metaplots for chromatin surrounding genes or TE insertion sites
For DNA methylation, the context-specific levels of DNA methylation for each 100-bp bin across the genome were utilized. For chromatin accessibility, the normalized (reads per million) values for the heavy and light digest were calculated for each 100-bp bin and the ratio (heavy/light) was used as a measure of relative accessibility. For each annotated attribute (gene, TIR, LTR), bedtools closest was used to calculate the distance of each 100-bp bin to that attribute. Relative distance was then determined for the 100-bp bins within the attribute (normalized to a 1,000-bp window). All 100-bp bin data for 1 kb upstream and downstream of the attribute were retained and data points were averaged for every 100-bp window; these values were then plotted across the genomic region relative to the gene, TIR or LTR attribute. Metaplots of chromatin features surrounding potential Ds or Mu insertion sites were performed by selecting the 100-bp bins upstream and downstream (10 kb) of each insertion site using bedtools. Data points were averaged for each 100-bp window across the genomic space relative to the insertion site (indicated by x-axis zero).
Analysis of chromatin at edges of TIR TE families
The DNA methylation and MNase data for 100-bp bins were compared with the TIR annotation gff using bedtools closest to identify the 100-bp bins within 4 kb of each TIR element. Only 100-bp bins that are closest to a single TIR element are used for analysis of chromatin properties at the edge of TIR elements. The TE family ID was identified and for each family (only those families with 10+ members were used to generate the figure; see Supplementary Table 8 for details on these families), the average level of DNA methylation or chromatin accessibility was calculated for each 100-bp window with respect to the TE edge. Mu and Ds data were then added for each DNA methylation context and MNase. The heatmap was then generated using a Euclidean clustering system focusing on the CG and CHG methylation levels. Once the family order was determined, a separate scale was used to generate each context of methylation and MNase to view the trend within each (CG/CHG = 0–1, CHH = 0–0.15, MNase = 0–4).
Reporting Summary
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The W22v2 genome sequence is available at NCBI under the accession GCA_001644905.2. The W22 annotations are available at MaizeGDB.org. There are also RNA-seq (SRR1986376), whole-genome bisulfite (SRX3136383) and MNase (SRP118121) data sets available at NCBI SRA.
References
Brink, R. A., Styles, E. D. & Axtell, J. D. Paramutation: directed genetic change. Paramutation occurs in somatic cells and heritably alters the functional state of a locus. Science 159, 161–170 (1968).
Mertz, E. T., Bates, L. S. & Nelson, O. E. Mutant gene that changes protein composition and increases lysine content of maize endosperm. Science 145, 279–280 (1964).
Kermicle, J. L. Dependence of the R-mottled aleurone phenotype in maize on mode of sexual transmission. Genetics 66, 69–85 (1970).
Fedoroff, N. V., Furtek, D. B. & Nelson, O. E. Cloning of the bronze locus in maize by a simple and generalizable procedure using the transposable controlling element Activator (Ac). Proc. Natl Acad. Sci. USA 81, 3825–3829 (1984).
Chen, J., Greenblatt, I. M. & Dellaporta, S. L. Transposition of Ac from the P locus of maize into unreplicated chromosomal sites. Genetics 117, 109–116 (1987).
Dooner, H., English, J., Ralston, E. & Weck, E. A single genetic unit specifies two transposition functions in the maize element activator. Science 234, 210–211 (1986).
McCarty, D. R. et al. Steady-state transposon mutagenesis in inbred maize. Plant J. 44, 52–61 (2005).
McCarty, D. R. et al. Mu-seq: sequence-based mapping and identification of transposon induced mutations. PLoS One 8, e77172 (2013).
Liu, P., McCarty, D. R. & Koch, K. E. Transposon mutagenesis and analysis of mutants in UniformMu maize (Zea mays). Curr. Protoc. Plant Biol. 1, 451–465.
Brutnell, T. P. & Conrad, L. J. Transposon tagging using Activator (Ac) in maize. Methods Mol. Biol. 236, 157–176 (2003).
Kolkman, J. M. et al. Distribution of Activator (Ac) throughout the maize genome for use in regional mutagenesis. Genetics 169, 981–995 (2005).
Li, Y., Segal, G., Wang, Q. & Dooner, H. K. Gene tagging with engineered Ds elements in maize. Methods Mol. Biol. 1057, 83–99 (2013).
Vollbrecht, E. et al. Genome-wide distribution of transposed Dissociation elements in maize. Plant Cell 22, 1667–1685 (2010).
Springer, N. M. et al. Maize inbreds exhibit high levels of copy number variation (CNV) and presence/absence variation (PAV) in genome content. PLoS Genet. 5, e1000734 (2009).
Lai, J. et al. Genome-wide patterns of genetic variation among elite maize inbred lines. Nat. Genet. 42, 1027–1030 (2010).
Fu, H. & Dooner, H. K. Intraspecific violation of genetic colinearity and its implications in maize. Proc. Natl Acad. Sci. USA 99, 9573–9578 (2002).
Hirsch, C. N. et al. Insights into the maize pan-genome and pan-transcriptome. Plant Cell 26, 121–135 (2014).
Jiao, Y. et al. Improved maize reference genome with single-molecule technologies. Nature 546, 524–527 (2017).
Hirsch, C. N. et al. Draft assembly of elite inbred line PH207 provides insights into genomic and transcriptome diversity in maize. Plant Cell 28, 2700–2714 (2016).
Zheng, G. X. Y. et al. Haplotyping germline and cancer genomes with high-throughput linked-read sequencing. Nat. Biotechnol. 34, 303–311 (2016).
Luo, M.-C. et al. Optical nano-mapping and analysis of plant genomes. Methods Mol. Biol. 1429, 103–117 (2016).
Lu, F. et al. High-resolution genetic mapping of maize pan-genome sequence anchors. Nat. Commun. 6, 6914 (2015).
Dooner, H. K. & He, L. Maize genome structure variation: interplay between retrotransposon polymorphisms and genic recombination. Plant Cell 20, 249–258 (2008).
Simão, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V. & Zdobnov, E. M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212 (2015).
Swanson-Wagner, R. A. et al. Pervasive gene content variation and copy number variation in maize and its undomesticated progenitor. Genome Res. 20, 1689–1699 (2010).
McCormick, R. F. et al. The Sorghum bicolor reference genome: improved assembly, gene annotations, a transcriptome atlas, and signatures of genome organization. Plant J. 93, 338–354 (2018).
Mei, W. et al. A comprehensive analysis of alternative splicing in paleopolyploid maize. Front. Plant Sci. 8, 694 (2017).
Schnable, P. S. et al. The B73 maize genome: complexity, diversity, and dynamics. Science 326, 1112–1115 (2009).
Dietrich, C. R. et al. Maize Mu transposons are targeted to the 5ʹ untranslated region of the gl8 gene and sequences flanking Mu target-site duplications exhibit nonrandom nucleotide composition throughout the genome. Genetics 160, 697–716 (2002).
Liu, S. et al. Mu transposon insertion sites and meiotic recombination events co-localize with epigenetic marks for open chromatin across the maize genome. PLoS Genet. 5, e1000733 (2009).
Gent, J. I. et al. CHH islands: de novo DNA methylation in near-gene chromatin regulation in maize. Genome Res. 23, 628–637 (2013).
Regulski, M. et al. The maize methylome influences mRNA splice sites and reveals widespread paramutation-like switches guided by small RNA. Genome Res. 23, 1651–1662 (2013).
Li, Q. et al. Examining the causes and consequences of context-specific differential DNA methylation in maize. Plant Physiol. 168, 1262–1274 (2015).
Sultana, T., Zamborlini, A., Cristofari, G. & Lesage, P. Integration site selection by retroviruses and transposable elements in eukaryotes. Nat. Rev. Genet. 18, 292–308 (2017).
Nannas, N. J. & Dawe, R. K. Genetic and genomic toolbox of Zea mays. Genetics 199, 655–669 (2015).
Messing, J. & Dooner, H. Organization and variability of the maize genome. Curr. Opin. Plant Biol. 9, 157–163 (2006).
Chia, J. M. et al. Maize HapMap2 identifies extant variation from a genome in flux. Nat. Genet. 44, 803–807 (2012).
Altpeter, F. et al. Advancing crop transformation in the era of genome editing. Plant Cell 28, 1510–1520 (2016).
Studer, A., Zhao, Q., Ross-Ibarra, J. & Doebley, J. Identification of a functional transposon insertion in the maize domestication gene tb1. Nat. Genet. 43, 1160–1163 (2011).
Yang, Q. et al. CACTA-like transposable element in ZmCCT attenuated photoperiod sensitivity and accelerated the postdomestication spread of maize. Proc. Natl Acad. Sci. USA 110, 16969–16974 (2013).
Castelletti, S., Tuberosa, R., Pindo, M., & Salvi, S. A MITE transposon insertion is associated with differential methylation at the maize flowering time QTL Vgt1. G3 4, 805–812 (2014).
Zhang, M. et al. Preparation of megabase-sized DNA from a variety of organisms using the nuclei method for advanced genomics research. Nat. Protoc. 7, 467–478 (2012).
Avni, R. et al. Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science 357, 93–97 (2017).
Campbell, M. S. et al. MAKER-P: a tool kit for the rapid creation, management, and quality control of plant genome annotations. Plant Physiol. 164, 513–524 (2014).
Law, M. et al. Automated update, revision, and quality control of the maize genome annotations using MAKER-P improves the B73 RefGen_v3 gene models and identifies new genes. Plant Physiol. 167, 25–39 (2015).
Wang, B. et al. Unveiling the complexity of the maize transcriptome by single-molecule long-read sequencing. Nat. Commun. 7, 11708 (2016).
Fu, L., Niu, B., Zhu, Z., Wu, S. & Li, W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152 (2012).
Martin, J. A. et al. A near complete snapshot of the Zea mays seedling transcriptome revealed from ultra-deep sequencing. Sci. Rep. 4, 4519 (2014).
Tello-Ruiz, M. K. et al. Gramene 2016: comparative plant genomics and pathway resources. Nucleic Acids Res. 44, D1133–40 (2016).
Stanke, M. et al. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res. 34, W435–W439 (2006).
Salamov, A. A. & Solovyev, V. V. Ab initio gene finding in Drosophila genomic DNA. Genome Res. 10, 516–522 (2000).
Wicker, T. et al. A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 8, 973–982 (2007).
Ellinghaus, D., Kurtz, S. & Willhoeft, U. LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinformatics 9, 18 (2008).
Han, Y., Burnette, J. M. 3rd & Wessler, S. R. TARGeT: a web-based pipeline for retrieving and characterizing gene and transposable element families from genomic sequences. Nucleic Acids Res. 37, e78 (2009).
Ye, C., Ji, G. & Liang, C. detectMITE: A novel approach to detect miniature inverted repeat transposable elements in genomes. Sci. Rep. 6, 19688 (2016).
Wenke, T. et al. Targeted identification of short interspersed nuclear element families shows their widespread existence and extreme heterogeneity in plant genomes. Plant Cell 23, 3117–3128 (2011).
Tang, H. et al. SynFind: compiling syntenic regions across any set of genomes on demand. Genome Biol. Evol. 7, 3286–3298 (2015).
Schnable, J. C. Genome evolution in maize: from genomes back to genes. Annu. Rev. Plant Biol. 66, 329–343 (2015).
Mei, W., Boatwright, L., Feng, G., Schnable, J. C. & Barbazuk, W. B. Evolutionarily conserved alternative splicing across monocots. Genetics 207, 465–480 (2017).
Acknowledgements
This work was supported by grants from the following: National Science Foundation NSF-MCB 0920218 to H.K.D.; NSF-MCB 1025976 to K.E.K.; NSF-IOS 1031416 to A.M.S.; NSF-IOS 1237931 to N.M.S.; NSF-IOS 1116561 to D.R.M. and K.E.K.; NSF-IOS 1238014 to E.S.B.; NSF-IOS 1314143 to T.P.B. and E.V.; NSF IOS-1339237 and IOS-1139329 to G.J.; NSF MCB-1412218 to A.M.S. and W.B.B.; NSF IOS-1444514 to R.K.D.; NSF-IOS 1521100 to J.-C.G., K.E.K. and D.R.M.; NSF-IOS 1547787 to W.B.B.; NSF Graduate Research Fellowship and University of Florida Graduate Student Fellowship to C.G.; NSF Graduate Research Fellowship DGE-0802270 to C.G.; the United States Department of Agriculture (USDA) in kind to C.M.A., E.K.S.C., M.R.W. and J.L.P.; USDA-ARS 6036-21000-011-00D to C.T.H.; USDA-NIFA 2016-67013-24747 to N.M.S.; USDA-NIFA 2011-67003-30215 to D.R.M. and K.E.K.; USDA-NIFA 2011-67013-30082 to D.R.M. and M.S.; USDA-ARS and the NSF-IOS Plant Genome 1238014 to E.S.B.; the Enterprise Rent-A-Car Institute for Renewable Fuels at the Donald Danforth Plant Science Center to T.P.B.; and the Vasil-Monsanto Endowment to A.M.S. The use of trade names, commercial products or corporations in this publication is for the information and convenience of the reader and does not imply an official recommendation, endorsement or approval by the US Department of Agriculture or the Agricultural Research Service for any product or service to the exclusion of others that may be suitable. USDA is an equal opportunity provider and employer.
Author information
Authors and Affiliations
Contributions
K.R.A., F.B., H.K.D., D.M.-J., T.K., J.S. and J.T.V. provided or isolated biological materials; O.B., K.B., G.B.-Z., G.K., G.R., D.S.-T. and I.S. performed the genome assembly; O.B., K.B., R.B., G.B.-Z., E.S.B., R.K.D., L.H.D., C.D., G.K., F.L., G.R., D.S.-T., E.V., J.S. and I.S. performed quality assessments and assembly validation; M.S.C., G.J., Y.J., T.G.K., J.C.S., M.C.S., D.W., S.W. and M.R.W. annotated the genome; C.M.A., E.K.S.C., K.A.E. and J.L.P. developed visualization tools; S.N.A., F.B., W.B.B., J.-C.G., R.D., C.G., K.E.K., T.K., D.M.-J., W.M., D.R.M., A.M.S. and M.S. performed transcriptome analyses; S.N.A., K.R.A., C.D., C.T.H., K.E.K., D.R.M., J.M.N., M.C.S., E.V., K.W. and W.X. mapped and characterized TE distribution; H.W.B., Q.L., J.M.N. and D.L.V. characterized epigenetic profiles; and T.P.B., P.C., H.K.D. and N.M.S. contributed to experimental design and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
Several authors are employed by companies that provide genome sequencing and assembly support. O.B., K.B., G.B.-Z., P.C., G.K., G.R., D.S.-T. and I.S. are employed by NRGene and J.S. is employed by BioNano Genomics.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Comparison of the completeness of the B73v4 and W22v2 genome annotations.
The completeness of the genome assemblies and genome annotations was assessed by benchmarking a universal single-copy orthologous gene set (BUSCO) (Simão et al. 2015). The relative frequencies of complete single-copy genes, duplicated genes, fragmented genes and missing genes were very similar in the B73v4 and W22v2 genomes.
Supplementary Figure 2 Density plot of shared and unique alternative splicing of W22 compared to B73v4.
For the B73v4 alternative splicing events, we mapped the isoforms from B73v4 annotation to the W22 genome and identified the alternative splicing events. We classified the common alternative splicing events based on the coordinates of the alternative region relative to the W22v2 genome.
Supplementary Figure 3 Improved transcriptome analyses using the W22v2 genome.
a, RNA-seq data derived from endosperm tissue of W22 (SRA: SRR1986376) were aligned to both B73v4 and W22v2 using the same parameters. The percentage of mapped reads improves by mapping the data to W22. b, A set of 20,994 syntenic orthologous genes in B73 and W22 were identified and used for comparison of expression levels in alignments to B73 or W22. A comparison of expression level (reads per million, RPM) shows generally similar estimates in both genotypes with some genes that have differing expression estimates depending upon which genome was used for a reference. c–f, Differences in RPKM estimates can result from differences in annotation or mapping efficiency. c,d, Overlapping gene models in B73 result in all reads mapping to Zm00001d028756 (orange transcript) to be called ambiguous, while the corresponding gene in W22, Zm00004b001227 (orange transcript), has reads assigned. The adjacent gene, Zm00004b001228 in W22 and Zm00001d028757 in B73 (teal transcripts), has the same number of reads assigned to each reference, with a lower RPKM value reported in B73 owing to the longer gene model. e,f, The RPKM value for genes Zm00004b005439 in W22 and Zm00001d034453 in B73 is higher when mapping to W22 (e) than to B73 (f) owing to improved alignment to several regions of the gene (marked in yellow). Mapped reads are colored by strand: blue, forward; red, reverse.
Supplementary Figure 4 Comparison of TIR copy number in B73 and W22.
a, The proportion of TIR families in each of the superfamilies (Activator (DTA); CACTA (DTC), PIF/Harbinger (DTH), Mutator (DTM), Tourist (DTT)) was determined for all TIR families (blue) in the B73 and W22 genomes. The proportion of TIR families in these categories was then determined for B73-specific (orange) and W22-specific (gray) families. b, The copy number in each TIR TE family is shown for B73 and W22. Color indicates superfamilies. c, The relative copy number for each LTR TE family in B73 and W22 is shown. In d, only families with <500 copies are shown. e, Boxplot of the percent identity of LTR sequences for LTR retrotransposons, demonstrating that elements in B73-specific families with at least five members (orange) are younger than members of shared families (blue) and elements in B73-specific families with fewer than five members (red). Line, median; box limits, first and third quartiles; whiskers, furthest point within 1.5 * IQR; points, outliers.
Supplementary Figure 5 Profiles of CHH methylation surrounding sites targeted by Ds (solid lines) or Mu (dashed lines).
The level of CHH methylation is shown for the flanking regions (up to 10 kb) near Ds or Mu insertion sites.
Supplementary Figure 6 Dendrogram analysis (unrooted tree) of terpene synthases in B73 and W22.
A set of 81 amino acid sequences (42 from B73 in blue and 39 from W22 in black) were used to generate a tree based on MUSCLE protein alignment by using the Maximum Likelihood method and a previously described substitution model. Bootstrap values (n = 1,000 replicates) are shown next to each node. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. All positions with less than 80% site coverage were eliminated. Evolutionary analyses were conducted in MEGA6. Asterisks are used to mark likely non-functional genes.
Supplementary Figure 7 Sequence conservation among Mutator transposable elements.
a, Diagram of a Mutator transposable element. The highly conserved terminal inverted repeats (TIRs) can be used to identify Mu elements and to categorize them based on phylogenetic groups. b, Alignment of consensus sequences for each of the seven Mu TIR groups. Highlighted nucleotides indicate disagreements with the overall consensus sequence. The terminal 20 positions are the most conserved across TIR groups. The predicted transposase-binding site (from position 34 to 68) is also highly conserved (57%) across the seven groups.
Supplementary Figure 8 Analysis of synteny between Mutator transposons identified in W22 and B73, including intact elements and orphan TIRs.
Of the 386 Mu elements or orphan TIRs examined, 133 were present in both the W22 and B73 genomes.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8, Supplementary Tables 1–9 and Supplementary Note
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Springer, N.M., Anderson, S.N., Andorf, C.M. et al. The maize W22 genome provides a foundation for functional genomics and transposon biology. Nat Genet 50, 1282–1288 (2018). https://doi.org/10.1038/s41588-018-0158-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-018-0158-0
This article is cited by
-
Plant pangenomes for crop improvement, biodiversity and evolution
Nature Reviews Genetics (2024)
-
A potential candidate gene associated with the angles of the ear leaf and the second leaf above the ear leaf in maize
BMC Plant Biology (2023)
-
The terpene synthase gene family in maize – a clarification of existing community nomenclature
BMC Genomics (2023)
-
A complete telomere-to-telomere assembly of the maize genome
Nature Genetics (2023)
-
Gene mining and genomics-assisted breeding empowered by the pangenome of tea plant Camellia sinensis
Nature Plants (2023)