Abstract
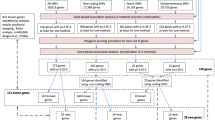
Neuroticism is an important risk factor for psychiatric traits, including depression1, anxiety2,3, and schizophrenia4,5,6. At the time of analysis, previous genome-wide association studies7,8,9,10,11,12 (GWAS) reported 16 genomic loci associated to neuroticism10,11,12. Here we conducted a large GWAS meta-analysis (n = 449,484) of neuroticism and identified 136 independent genome-wide significant loci (124 new at the time of analysis), which implicate 599 genes. Functional follow-up analyses showed enrichment in several brain regions and involvement of specific cell types, including dopaminergic neuroblasts (P = 3.49 × 10−8), medium spiny neurons (P = 4.23 × 10−8), and serotonergic neurons (P = 1.37 × 10−7). Gene set analyses implicated three specific pathways: neurogenesis (P = 4.43 × 10−9), behavioral response to cocaine processes (P = 1.84 × 10−7), and axon part (P = 5.26 × 10−8). We show that neuroticism’s genetic signal partly originates in two genetically distinguishable subclusters13 (‘depressed affect’ and ‘worry’), suggesting distinct causal mechanisms for subtypes of individuals. Mendelian randomization analysis showed unidirectional and bidirectional effects between neuroticism and multiple psychiatric traits. These results enhance neurobiological understanding of neuroticism and provide specific leads for functional follow-up experiments.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Kendler, K. S. & Myers, J. The genetic and environmental relationship between major depression and the five-factor model of personality. Psychol. Med. 40, 801–806 (2010).
Middeldorp, C. M. et al. in Biology of Personal and Individual Differences (ed. Canli, T.) Ch. 12, 251–272 (Guilford Press, New York and London, 2006).
Hettema, J. M., Neale, M. C., Myers, J. M., Prescott, C. A. & Kendler, K. S. A population-based twin study of the relationship between neuroticism and internalizing disorders. Am. J. Psychiatry 163, 857–864 (2006).
Hayes, J. F., Osborn, D. P. J., Lewis, G., Dalman, C. & Lundin, A. Association of late adolescent personality with risk for subsequent serious mental illness among men in a Swedish nationwide cohort study. JAMA Psychiatry 74, 703–711 (2017).
Smeland, O. B. et al. Identification of genetic loci shared between schizophrenia and the Big Five personality traits. Sci. Rep. 7, 2222 (2017).
Van Os, J. & Jones, P. B. Neuroticism as a risk factor for schizophrenia. Psychol. Med 31, 1129–1134 (2001).
Genetics of Personality Consortium. Meta-analysis of genome-wide association studies for neuroticism, and the polygenic association with major depressive disorder. JAMA Psychiatry 72, 642–650 (2015).
Terracciano, A. et al. Genome-wide association scan for five major dimensions of personality. Mol. Psychiatry 15, 647–656 (2010).
de Moor, M. H. M. et al. Meta-analysis of genome-wide association studies for personality. Mol. Psychiatry 17, 337–349 (2012).
Lo, M. T. et al. Genome-wide analyses for personality traits identify six genomic loci and show correlations with psychiatric disorders. Nat. Genet. 49, 152–156 (2017).
Smith, D. J. et al. Genome-wide analysis of over 106,000 individuals identifies 9 neuroticism-associated loci. Mol. Psychiatry 21, 1–9 (2016).
Okbay, A. et al. Genetic variants associated with subjective well-being, depressive symptoms and neuroticism identified through genome-wide analyses. Nat. Genet. 48, 624–633 (2016).
Nagel, M., Watanabe, K., Stringer, S., Posthuma, D. & van der Sluis, S. Item-level analyses reveal genetic heterogeneity in neuroticism. Nat. Commun. 9, 905 (2018).
Bycroft, C. et al. Genome-wide genetic data on ~500,000 UK Biobank participants. Preprint at bioRxiv https://doi.org/10.1101/166298 (2017).
Eriksson, N. et al. Web-based, participant-driven studies yield novel genetic associations for common traits. PLoS Genet. 6, e1000993 (2010).
Willer, C. J., Li, Y., Abecasis, G. R. & Overall, P. METAL: fast and efficient meta-analysis of genome-wide association scans. Bioinformatics 26, 2190–2191 (2010).
Skol, A. D., Scott, L. J., Abecasis, G. R. & Boehnke, M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat. Genet. 38, 209–213 (2006).
Bulik-Sullivan, B. K. et al. LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47, 291–295 (2015).
Yang, J. et al. Genomic inflation factors under polygenic inheritance. Eur. J. Hum. Genet. 19, 807–812 (2011).
Watanabe, K., Taskesen, E., van Bochoven, A. & Posthuma, D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 8, 1826 (2017).
Kircher, M. et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 46, 310–315 (2014).
Finucane, H. K. et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet. 47, 1228–1235 (2015).
Euesden, J., Lewis, C. M. & O’Reilly, P. F. PRSice: polygenic risk score software. Bioinformatics 31, 1466–1468 (2015).
Vilhjálmsson, B. J. et al. Modeling linkage disequilibrium increases accuracy of polygenic risk scores. Am. J. Hum. Genet. 97, 576–592 (2015).
de Leeuw, C. A., Mooij, J. M., Heskes, T. & Posthuma, D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput. Biol. 11, e1004219 (2015).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
GTEx Consortium. The genotype–tissue expression (GTEx) pilot analysis: multi-tissue gene regulation in humans. Science 348, 648–660 (2015).
Skene, N. G. et al. Genetic identification of brain cell types underlying schizophrenia. Nat. Genet. 50, 825–833 (2018).
Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium. A mega-analysis of genome-wide association studies for major depressive disorder. Mol. Psychiatry 18, 497–511 (2013).
Bulik-Sullivan, B. et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 47, 1236–1241 (2015).
Zhu, Z. et al. Causal associations between risk factors and common diseases inferred from GWAS summary data. Nat. Commun. 9, 224 (2018).
Griffith, M. et al. DGIdb: mining the druggable genome. Nat. Methods 10, 1209–1210 (2013).
Cotto, K. C. et al. DGIdb 3.0: a redesign and expansion of the drug–gene interaction database. Nucleic Acids Res. 46, D1068–D1073 (2017).
Luciano, M. et al. Association analysis in over 329,000 individuals identifies 116 independent variants influencing neuroticism. Nat. Genet. 50, 6–11 (2018).
Hyde, C. L. et al. Identification of 15 genetic loci associated with risk of major depression in individuals of European descent. Nat. Genet. 48, 1031–1036 (2016).
Eysenck, B. G., Eysenck, H. J. & Barrett, P. A revised version of the psychoticism scale. Pers. Individ. Dif. 6, 21–29 (1985).
John, O. P. & Srivastava, S. The Big Five trait taxonomy: history, measurement and theoretical perspectives. Handb. Personal. Theory Res. 2, 102–138 (1999).
Soto, C. J. & John, O. P. Ten facet scales for the Big Five Inventory: convergence with NEO PI-R facets, self-peer agreement and discriminant validity. J. Res. Pers. 43, 84–90 (2009).
Costa, P. & McCrae, R. R. Professional Manual: Revised NEO Personality Inventory (NEO-PI-R) and NEO Five-Factor-Inventory (NEO-FFI). (Psychological Assessment Resources, Odessa, FL, USA, 1992).
Auton, A. et al. A global reference for human genetic variation. Nature 526, 68–74 (2015).
Webb, B. T. et al. Molecular genetic influences on normative and problematic alcohol use in a population-based sample of college students. Front. Genet. 8, 30 (2017).
Abraham, G. & Inouye, M. Fast principal component analysis of large-scale genome-wide data. PLoS One 9, e93766 (2014).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Chang, C. C. et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4, 7 (2015).
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010).
Boyle, A. P. et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 22, 1790–1797 (2012).
Schmitt, A. D. et al. A compendium of chromatin contact maps reveals spatially active regions in the human genome. Cell Rep. 17, 2042–2059 (2016).
Roadmap Epigenomics Consortium. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015).
Croft, D. et al. The Reactome pathway knowledgebase. Nucleic Acids Res. 42, D472–D477 (2014).
Coleman, J. R. I. et al. Functional consequences of genetic loci associated with intelligence in a meta-analysis of 87,740 individuals. Mol. Psychiatry https://doi.org/10.1038/s41380-018-0040-6 (2018).
Acknowledgements
We would like to thank the participants, including the 23andMe customers who consented to participate in research, and the researchers who collected and contributed to the data. This work was funded by the Netherlands Organization for Scientific Research through the following grants: NWO Brain and Cognition 433-09-228 (D.P.), NWO MagW VIDI 452-12-014 (S.v.d.S.), NWO VICI 435-13-005 (D.P.) and 645-000-003) (D.P.). P.R.J. was funded by the Sophia Foundation for Scientific Research (SSWO, grant no. S14-27). J.H.-L. was funded by the Swedish Research Council (Vetenskapsrådet, award 2014-3863), StratNeuro, the Wellcome Trust (108726/Z/15/Z), and the Swedish Brain Foundation (Hjärnfonden). N.G.S. was supported by the Wellcome Trust (108726/Z/15/Z). J.B. was funded by the Swiss National Science Foundation. The work of H.T. was supported by a NWO–VICI grant (NWO-ZonMW 016.VICI.170.200). Analyses were carried out on the Genetic Cluster computer, which is financed by the Netherlands Scientific Organization (NWO award 480-05-003 to D.P.), VU University (Amsterdam, The Netherlands), and the Dutch Brain Foundation and is hosted by the Dutch National Computing and Networking Services, SurfSARA. This research has been conducted using the UK Biobank resource (application 16406).
Author information
Authors and Affiliations
Consortia
Contributions
S.v.d.S. and D.P. conceived the study; M.N. and P.R.J. performed the analyses; S.S. performed the quality control on the UKB data and wrote a pipeline to facilitate data processing; K.W. constructed the tool for biological annotation and ran the analyses; H.T. and T.W. read and commented on the pre-final version of the manuscript; A.R.H., C.A.d.L., J.E.S., and T.J.C.P. wrote part of the analysis pipeline and assisted in interpreting results; N.G.S., A.B.M.-M., S.L., and J.H.-L. provided single-cell RNA-seq data for mouse brain cell types; J.B. and P.F.S. performed the single-cell gene expression analysis; and M.N., P.R.J., S.v.d.S., and D.P. wrote the paper. All authors discussed the results and commented on the paper.
Corresponding author
Ethics declarations
Competing interests
J.H.-L. is a scientific advisor at Cartana and has received a grant from Roche. P.F. has received a grant from Lundbeck and is currently a member of the advisory committee. Over the last 3 years, P.F. has been on the scientific advisory board at Pfizer, received a consultation fee from Element Genomics, and received speaker reimbursement fees from Roche.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Text and Figures
Supplementary Note, Supplementary Results and Supplementary Figures 1–17
Rights and permissions
About this article
Cite this article
Nagel, M., Jansen, P.R., Stringer, S. et al. Meta-analysis of genome-wide association studies for neuroticism in 449,484 individuals identifies novel genetic loci and pathways. Nat Genet 50, 920–927 (2018). https://doi.org/10.1038/s41588-018-0151-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-018-0151-7
This article is cited by
-
PASTRY: achieving balanced power for detecting risk and protective minor alleles in meta-analysis of association studies with overlapping subjects
BMC Bioinformatics (2024)
-
Olfactory genes affect major depression in highly educated, emotionally stable, lean women: a bridge between animal models and precision medicine
Translational Psychiatry (2024)
-
Patterns of stressful life events and polygenic scores for five mental disorders and neuroticism among adults with depression
Molecular Psychiatry (2024)
-
A concerted neuron–astrocyte program declines in ageing and schizophrenia
Nature (2024)
-
Key subphenotypes of bipolar disorder are differentially associated with polygenic liabilities for bipolar disorder, schizophrenia, and major depressive disorder
Molecular Psychiatry (2024)