Abstract
Enhancers act to regulate cell-type-specific gene expression by facilitating the transcription of target genes. In mammalian cells, active or primed enhancers are commonly marked by monomethylation of histone H3 at lysine 4 (H3K4me1) in a cell-type-specific manner. Whether and how this histone modification regulates enhancer-dependent transcription programs in mammals is unclear. In this study, we conducted SILAC mass spectrometry experiments with mononucleosomes and identified multiple H3K4me1-associated proteins, including many involved in chromatin remodeling. We demonstrate that H3K4me1 augments association of the chromatin-remodeling complex BAF to enhancers in vivo and that, in vitro, H3K4me1-marked nucleosomes are more efficiently remodeled by the BAF complex. Crystal structures of the BAF component BAF45C indicate that monomethylation, but not trimethylation, is accommodated by BAF45C’s H3K4-binding site. Our results suggest that H3K4me1 has an active role at enhancers by facilitating binding of the BAF complex and possibly other chromatin regulators.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Hardison, R. C. & Taylor, J. Genomic approaches towards finding cis-regulatory modules in animals. Nat. Rev. Genet. 13, 469–483 (2012).
Heintzman, N. D. et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 39, 311–318 (2007).
Creyghton, M. P. et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA 107, 21931–21936 (2010).
Heintzman, N. D. et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459, 108–112 (2009).
Heinz, S., Romanoski, C. E., Benner, C. & Glass, C. K. The selection and function of cell type-specific enhancers. Nat. Rev. Mol. Cell Biol. 16, 144–154 (2015).
Rada-Iglesias, A. et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470, 279–283 (2011).
Taylor, G. C., Eskeland, R., Hekimoglu-Balkan, B., Pradeepa, M. M. & Bickmore, W. A. H4K16 acetylation marks active genes and enhancers of embryonic stem cells, but does not alter chromatin compaction. Genome Res. 23, 2053–2065 (2013).
Pradeepa, M. M. et al. Histone H3 globular domain acetylation identifies a new class of enhancers. Nat. Genet. 48, 681–686 (2016).
Calo, E. & Wysocka, J. Modification of enhancer chromatin: what, how, and why? Mol. Cell 49, 825–837 (2013).
Musselman, C. A., Lalonde, M. E., Côté, J. & Kutateladze, T. G. Perceiving the epigenetic landscape through histone readers. Nat. Struct. Mol. Biol. 19, 1218–1227 (2012).
Seet, B. T., Dikic, I., Zhou, M. M. & Pawson, T. Reading protein modifications with interaction domains. Nat. Rev. Mol. Cell Biol. 7, 473–483 (2006).
Smith, E. & Shilatifard, A. The chromatin signaling pathway: diverse mechanisms of recruitment of histone-modifying enzymes and varied biological outcomes. Mol. Cell 40, 689–701 (2010).
Strahl, B. D. & Allis, C. D. The language of covalent histone modifications. Nature 403, 41–45 (2000).
Vermeulen, M. et al. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell 131, 58–69 (2007).
Hu, D. et al. The MLL3/MLL4 branches of the COMPASS family function as major histone H3K4 monomethylases at enhancers. Mol. Cell. Biol. 33, 4745–4754 (2013).
Lee, J. E. et al. H3K4 mono- and di-methyltransferase MLL4 is required for enhancer activation during cell differentiation. eLife 2, e01503 (2013).
Wang, C. et al. Enhancer priming by H3K4 methyltransferase MLL4 controls cell fate transition. Proc. Natl. Acad. Sci. USA 113, 11871–11876 (2016).
Outchkourov, N. S. et al. Balancing of histone H3K4 methylation states by the Kdm5c/SMCX histone demethylase modulates promoter and enhancer function. Cell Rep. 3, 1071–1079 (2013).
Cheng, J. et al. A role for H3K4 monomethylation in gene repression and partitioning of chromatin readers. Mol. Cell 53, 979–992 (2014).
Bartke, T. et al. Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell 143, 470–484 (2010).
Vermeulen, M. et al. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell 142, 967–980 (2010).
Carruthers, L. M., Tse, C., Walker, K. P. III & Hansen, J. C. Assembly of defined nucleosomal and chromatin arrays from pure components. Methods Enzymol. 304, 19–35 (1999).
Luger, K., Rechsteiner, T. J. & Richmond, T. J. Expression and purification of recombinant histones and nucleosome reconstitution. Methods Mol. Biol. 119, 1–16 (1999).
Luger, K., Rechsteiner, T. J. & Richmond, T. J. Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 304, 3–19 (1999).
Simon, M. D. et al. The site-specific installation of methyl-lysine analogs into recombinant histones. Cell 128, 1003–1012 (2007).
Engelen, E. et al. Proteins that bind regulatory regions identified by histone modification chromatin immunoprecipitations and mass spectrometry. Nat. Commun. 6, 7155 (2015).
Kagey, M. H. et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467, 430–435 (2010).
Dreveny, I. et al. The double PHD finger domain of MOZ/MYST3 induces α-helical structure of the histone H3 tail to facilitate acetylation and methylation sampling and modification. Nucleic Acids Res 42, 822–835 (2014).
Yue, F. et al. A comparative encyclopedia of DNA elements in the mouse genome. Nature 515, 355–364 (2014).
Li, Y. et al. CRISPR reveals a distal super-enhancer required for Sox2 expression in mouse embryonic stem cells. PLoS One 9, e114485 (2014).
Dorighi, K. M. et al. Mll3 and Mll4 facilitate enhancer RNA synthesis and transcription from promoters independently of H3K4 monomethylation. Mol. Cell 66, 568–576 (2017).
Phelan, M. L., Sif, S., Narlikar, G. J. & Kingston, R. E. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol. Cell 3, 247–253 (1999).
Zeng, L. et al. Mechanism and regulation of acetylated histone binding by the tandem PHD finger of DPF3b. Nature 466, 258–262 (2010).
Li, W., Zhao, A., Tempel, W., Loppnau, P. & Liu, Y. Crystal structure of DPF3b in complex with an acetylated histone peptide. J. Struct. Biol. 195, 365–372 (2016).
Xiong, X. et al. Selective recognition of histone crotonylation by double PHD fingers of MOZ and DPF2. Nat. Chem. Biol. 12, 1111–1118 (2016).
Ho, L. & Crabtree, G. R. Chromatin remodelling during development. Nature 463, 474–484 (2010).
Rickels, R. et al. Histone H3K4 monomethylation catalyzed by Trr and mammalian COMPASS-like proteins at enhancers is dispensable for development and viability. Nat. Genet. 49, 1647–1653 (2017).
Dyer, P. N. et al. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 375, 23–44 (2004).
Carey, M. F., Peterson, C. L. & Smale, S. T. Dignam and Roeder nuclear extract preparation. Cold Spring Harb. Protoc. 2009, pdb.prot5330 (2009).
Dignam, J. D., Martin, P. L., Shastry, B. S. & Roeder, R. G. Eukaryotic gene transcription with purified components. Methods Enzymol. 101, 582–598 (1983).
Kapust, R. B. & Waugh, D. S. Escherichia coli maltose-binding protein is uncommonly effective at promoting the solubility of polypeptides to which it is fused. Protein Sci. 8, 1668–1674 (1999).
Carey, M. F., Peterson, C. L. & Smale, S. T. In vivo DNase I, MNase, and restriction enzyme footprinting via ligation-mediated polymerase chain reaction (LM-PCR). Cold Spring Harb. Protoc. 2009, pdb.prot5277 (2009).
Albuquerque, C. P. et al. A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol. Cell. Proteomics 7, 1389–1396 (2008).
Chen, S. H., Albuquerque, C. P., Liang, J., Suhandynata, R. T. & Zhou, H. A proteome-wide analysis of kinase–substrate network in the DNA damage response. J. Biol. Chem. 285, 12803–12812 (2010).
Lowary, P. T. & Widom, J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 276, 19–42 (1998).
Cao, X. J., Arnaudo, A. M. & Garcia, B. A. Large-scale global identification of protein lysine methylation in vivo. Epigenetics 8, 477–485 (2013).
Yun, M., Ruan, C., Huh, J. W. & Li, B. Reconstitution of modified chromatin templates for in vitro functional assays. Methods Mol. Biol. 833, 237–253 (2012).
Kim, T. H. et al. Direct isolation and identification of promoters in the human genome. Genome Res. 15, 830–839 (2005).
Li, Z. et al. A global transcriptional regulatory role for c-Myc in Burkitt’s lymphoma cells. Proc. Natl. Acad. Sci. USA 100, 8164–8169 (2003).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Kabsch, W. Xds Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
Evans, P. R. & Murshudov, G. N. How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 69, 1204–1214 (2013).
Winn, M. D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011).
Terwilliger, T. C. et al. Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallogr. D Biol. Crystallogr. 65, 582–601 (2009).
McCoy, A. J., Storoni, L. C. & Read, R. J. Simple algorithm for a maximum-likelihood SAD function. Acta Crystallogr. D Biol. Crystallogr. 60, 1220–1228 (2004).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Terwilliger, T. C. Maximum-likelihood density modification. Acta Crystallogr. D Biol. Crystallogr. 56, 965–972 (2000).
Terwilliger, T. C. Automated main-chain model building by template matching and iterative fragment extension. Acta Crystallogr. D Biol. Crystallogr. 59, 38–44 (2003).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Afonine, P. V. et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 (2012).
Acknowledgements
The authors thank M. Carey (UCLA) for WT and mutant histone constructs, S. Kuan and B. Li for processing of ChIP–seq samples, J. Liang and G. Hon for help and advice in SILAC mass spectrometry analysis, J. Wysocka and K. Dorighi (Stanford School of Medicine) for sharing the KMT2C/D dCD mESC line, and I. Jung for advice on ChIP–seq data analysis. We also thank T. Gahman (Ludwig Institute for Cancer Research, LICR) for arranging for peptide synthesis and A. Bobkov for assistance with isothermal titration calorimetry. The research was supported in part by 5R01GM115961. C.P.A. and H.Z. were supported by funding from LICR and NIH GM116897. A.K.S., K.D.C., H.Z. and B.R. received funding and salary support from LICR. W.W. and D.W. were supported by funding from NIH GM102362. A.L. was supported by NIH Training Grant 5T32CA009523.
Author information
Authors and Affiliations
Contributions
A.L. and B.R. conceived the study and prepared the manuscript. A.L. designed and carried out the SILAC experiments, nucleosome pulldown experiments, and ChIP–seq experiments and prepared the manuscript. H.H. performed H3K4me2 ChIP–seq analysis and all experiments with the dCD cell lines. A.Y.L. prepared sequencing libraries. C.P.A. ran the mass spectrometry samples in the laboratory of H.Z. and provided expertise in mass spectrometry analysis. H.H. performed ChIP–seq data analysis. C.W. and K.G. provided KMT2C/D DKO mESCs and shared expertise and data. W.W. and D.W. designed and executed the remodeling assays. A.K.S. designed and supplied H3 tail peptides and, along with J.E.H., provided advice on their use in biochemical studies. N.S. purified BAF45C, performed H3 tail peptide binding measurements, and determined crystal structures under the direction of K.D.C.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
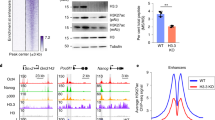
Supplementary Figure 1 The genomic distribution of candidate H3K4me1 binders correlates with enhancers in mESCs.
a, ChIP–qPCR with H3K4me1, H3K4me3, and H3K27ac antibodies; primers were designed for previously validated mESC enhancers E147, E132, E110, E151, and E8 and for negative-control region N9. Data are shown as means ± s.d.; n = 3 biological replicates. b, Left, browser shot of the E110 enhancer region. Right, browser shot of the N9 negative-control region. c, Browser shot of CR binding to the Nanog enhancer region. The left box highlights the active enhancer, and the right box highlights the poised enhancer. d, Browser shot of CR binding to the mESC-specific miR290 super-enhancer. e, Bar plots showing the fraction of enhancers occupied by CRs for active enhancers (n = 13,811) versus poised enhancers (n = 28,008); related to Fig. 2d. ChIP–seq experiments were repeated at least twice with each antibody.
Supplementary Figure 2 ChIP analysis of CR and histone modifications in WT and KMT2C/D DKO cells.
a, Loss of CR binding at the miR290 super-enhancer in KMT2C/D DKO mESCs. b, KMT2C/D-dependent site. c, KMT2C/D-independent site. Each experiment was repeated at least twice. d, Top, pie chart showing the fraction of H3K4me2 peaks in KMT2C/D DKO mESCs according to KMT2C/D-dependent and KMT2C/D-independent patterns. Bottom, 2 × 2 table of the relationship with enhancer regions according to KMT2C/D-dependent and KMT2C/D-independent H3K4me2 peak regions. e, ChIP–qPCR analysis of the CRs listed at the miR290-295 super-enhancer, the Nanog ESC enhancer, and a normal enhancer. The fold difference in binding for WT versus DKO cells was calculated from the average of three replicates.
Supplementary Figure 3 Depletion of H3K4me1 is associated with reduced binding of BAF components in KMT2C/D catalytically null (dCD) cells.
a, Left, predicted domains of mouse DPF2 and DPF3. Right, corresponding aligned sequence with the PHD domains in green. b, Distribution of distal H3K4me1, H3K4me2, and H3K4me3 regions in dCD cells as compared to WT cells. c, Heat map of ChIP–seq signal for DPF2, H3K27ac, H3K4me2, and H3K4me3 at distal H3K4me1 regions. Regions are sorted by strength of H3K4me1 signal. d, Aggregate plots showing average DPF2, H3K27ac, H3K4me2, and H3K4me3 ChIP signal in WT and dCD cells over the same regions in c.
Supplementary Figure 4 Purified BAF complex binding and remodeling activity.
a, Silver staining of purified FLAG-BAF complex. Subunits and sizes are indicated on the left, and ladder is on the right. A representative gel is shown for four replicate preparations. b, FLAG-BAF binding to methylated peptides. Binding was assayed by western blotting with FLAG antibody; the assay was repeated twice. c, Polyacrylamide gels showing the n = 4 replicate assays of nucleosome remodeling quantified in Fig. 4c.
Supplementary Figure 5 Binding of BAF45C to histone H3 tail peptides and structure of the BAF45C–H3 tail complex.
a, Isothermal titration calorimetry data showing interaction of the H31–18 H4K4me0/H3K14ac peptide (injectant) with purified DPF3 PHD1–PHD2 region. The K value of 1.28 × 105 ± 7.38 × 103 M–1 corresponds to a Kd of 7.8 ± 0.5 μM. b, Binding to H31–18 H4K4me1/H3K14ac. The K value of 4.90 × 104 ± 5.48 × 103 M–1 corresponds to a Kd of 20.4 ± 2.3 μM. c, Binding to H31–18 H4K4me3/H3K14ac. The K value of 8.72 × 103 ± 5.63 × 103 M–1 corresponds to a Kd of 115 ± 128 μM. d, Experimental electron density calculated from a single-wavelength Zn SAD dataset. e, Refined 2Fo – Fc electron density for the BAF45C–H3K4me0 complex. f, Refined 2Fo – Fc electron density for the DPF3–H3K4me1 complex. The experiment was performed once.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6.
Supplementary Table 1
Results from SILAC experiments.
Supplementary tables 2–5
Supplementary Tables 2–5.
Rights and permissions
About this article
Cite this article
Local, A., Huang, H., Albuquerque, C.P. et al. Identification of H3K4me1-associated proteins at mammalian enhancers. Nat Genet 50, 73–82 (2018). https://doi.org/10.1038/s41588-017-0015-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-017-0015-6
This article is cited by
-
Interrogating epigenetic mechanisms with chemically customized chromatin
Nature Reviews Genetics (2024)
-
Identification of Regulatory Elements in Primary Sensory Neurons Involved in Trauma-Induced Neuropathic Pain
Molecular Neurobiology (2024)
-
Loss of MLL3/4 decouples enhancer H3K4 monomethylation, H3K27 acetylation, and gene activation during embryonic stem cell differentiation
Genome Biology (2023)
-
Impaired histone inheritance promotes tumor progression
Nature Communications (2023)
-
Cell-specific and shared regulatory elements control a multigene locus active in mammary and salivary glands
Nature Communications (2023)