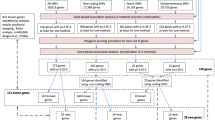
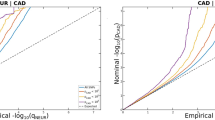
Abstract
Neuroticism is a relatively stable personality trait characterized by negative emotionality (for example, worry and guilt)1; heritability estimated from twin studies ranges from 30 to 50%2, and SNP-based heritability ranges from 6 to 15%3,4,5,6. Increased neuroticism is associated with poorer mental and physical health7,8, translating to high economic burden9. Genome-wide association studies (GWAS) of neuroticism have identified up to 11 associated genetic loci3,4. Here we report 116 significant independent loci from a GWAS of neuroticism in 329,821 UK Biobank participants; 15 of these loci replicated at P < 0.00045 in an unrelated cohort (N = 122,867). Genetic signals were enriched in neuronal genesis and differentiation pathways, and substantial genetic correlations were found between neuroticism and depressive symptoms (rg = 0.82, standard error (s.e.) = 0.03), major depressive disorder (MDD; rg = 0.69, s.e. = 0.07) and subjective well-being (rg = –0.68, s.e. = 0.03) alongside other mental health traits. These discoveries significantly advance understanding of neuroticism and its association with MDD.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Change history
29 January 2019
In the version of this article initially published, in Table 2, the descriptions of pathways and definitions in the first and last columns did not correctly correspond to the values in the other columns. The error has been corrected in the HTML and PDF versions of the article.
References
Matthews, G., Deary, I. J. & Whiteman, M. C. Personality Traits (Cambridge University Press, Cambridge, UK, 2009).
Vukasovic, T. & Bratko, D. Heritability of personality: a meta-analysis of behavior genetic studies. Psychol. Bull. 141, 769–785 (2015).
Smith, D. J. et al. Genome-wide analysis of over 106 000 individuals identifies 9 neuroticism-associated loci. Mol. Psychiatry 21, 749–757 (2016).
Okbay, A. et al. Genetic variants associated with subjective well-being, depressive symptoms, and neuroticism identified through genome-wide analyses. Nat. Genet. 48, 624–633 (2016).
Power, R. A. & Pluess, M. Heritability estimates of the Big Five personality traits based on common genetic variants. Transl. Psychiatry 5, e604 (2015).
Vinkhuyzen, A. A. et al. Common SNPs explain some of the variation in the personality dimensions of neuroticism and extraversion. Transl. Psychiatry 2, e102 (2012).
Kubzansky, L. D., Martin, L. T. & Buka, S. L. Early manifestations of personality and adult health: a life course perspective. Health. Psychol. 28, 364–372 (2009).
Strickhouser, J. E., Zell, E. & Krizan, Z. Does personality predict health and well-being? A metasynthesis. Health. Psychol. 36, 797–810 (2017).
Cuijpers, P. et al. Economic costs of neuroticism: a population-based study. Arch. Gen. Psychiatry 67, 1086–1093 (2010).
Few, L. R. et al. Genetic variation in personality traits explains genetic overlap between borderline personality features and substance use disorders. Addiction 109, 2118–2127 (2014).
Kendler, K. S., Gatz, M., Gardner, C. O. & Pedersen, N. L. Personality and major depression: a Swedish longitudinal, population-based twin study. Arch. Gen. Psychiatry 63, 1113–1120 (2006).
Wray, N. R. & Sullivan, P. F. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Preprint at bioRxiv https://doi.org/10.1101/167577 (2017).
Sudlow, C. et al. UK Biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 12, e1001779 (2015).
Bycroft, C. et al. Genome-wide genetic data on ~500,000 UK Biobank participants. Preprint at bioRxiv https://doi.org/10.1101/166298 (2017).
Eysenck, S. B., Eysenck, H. J. & Barrett, P. A revised version of the psychoticism scale. Pers. Individ. Dif. 6, 21–29 (1985).
McCarthy, S. et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet. 48, 1279–1283 (2016).
Bulik-Sullivan, B. K. et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47, 291–295 (2015).
Cross-Disorder Group of the Psychiatric Genomics Consortium. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet 381, 1371–1379 (2013).
Sekar, A. et al. Schizophrenia risk from complex variation of complement component 4. Nature 530, 177–183 (2016).
Hegyi, H. GABBR1 has a HERV-W LTR in its regulatory region—a possible implication for schizophrenia. Biol. Direct 8, 5 (2013).
Wei, J. & Hemmings, G. P. TNXB locus may be a candidate gene predisposing to schizophrenia. Am. J. Med. Genet. B Neuropsychiatr. Genet. 125B, 43–49 (2004).
Lo, M.-T. et al. Genome-wide analyses for personality traits identify six genomic loci and show correlations with psychiatric disorders. Nat. Genet. 49, 152–156 (2017).
de Moor, M. H. et al. Meta-analysis of genome-wide association studies for neuroticism, and the polygenic association with major depressive disorder. JAMA Psychiatry 72, 642–650 (2015).
Hu, Y. et al. GWAS of 89,283 individuals identifies genetic variants associated with self-reporting of being a morning person. Nat. Commun. 7, 10448 (2016).
Duggan, K. A., Friedman, H. S., McDevitt, E. A. & Mednick, S. C. Personality and healthy sleep: the importance of conscientiousness and neuroticism. PLoS One 9, e90628 (2014).
Mi, H., Muruganujan, A. & Thomas, P. D. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 41, D377–D386 (2013).
Sulser, F. The role of CREB and other transcription factors in the pharmacotherapy and etiology of depression. Ann. Med. 34, 348–356 (2002).
Wang, H. et al. Forkhead box O transcription factors as possible mediators in the development of major depression. Neuropharmacology 99, 527–537 (2015).
Malan-Müller, S., Hemmings, S. M. J. & Seedat, S. Big effects of small RNAs: a review of microRNAs in anxiety. Mol. Neurobiol. 47, 726–739 (2013).
Dwivedi, Y. Emerging role of microRNAs in major depressive disorder: diagnosis and therapeutic implications. Dialogues Clin. Neurosci. 16, 43–61 (2014).
Ambrosini, A. et al. Possible involvement of the CACNA1E gene in migraine: a search for single nucleotide polymorphism in different clinical phenotypes. Headache 57, 1136–1144 (2017).
Schraa-Tam, C. K. L. et al. fMRI activities in the emotional cerebellum: a preference for negative stimuli and goal-directed behavior. Cerebellum 11, 233–245 (2012).
Schutter, D. J. L. G., Koolschijn, P. C. M. P., Peper, J. S. & Crone, E. A. The cerebellum link to neuroticism: a volumetric MRI association study in healthy volunteers. PLoS One 7, e37252 (2012).
Grinberg, M. et al. Mitochondrial carrier homolog 2 is a target of tBID in cells signaled to die by tumor necrosis factor α. Mol. Cell. Biol. 25, 4579–4590 (2005).
Locke, A. E. et al. Genetic studies of body mass index yield new insights for obesity biology. Nature 518, 197–206 (2015).
de Leeuw, C. A., Mooij, J. M., Heskes, T. & Posthuma, D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput. Biol. 11, e1004219 (2015).
Lucassen, P. J., Oomen, C. A., Schouten, M., Encinas, J. M. & Fitzsimons, C. P. in Adult Neurogenesis in the Hippocampus (ed. Canales, J. J.) 177–206 (Academic Press, San Diego, CA, 2016).
Schoenfeld, T. J. & Cameron, H. A. Adult neurogenesis and mental illness. Neuropsychopharmacology 40, 113–128 (2015).
Wray, N. R. et al. Anxiety and comorbid measures associated with PLXNA2. Arch. Gen. Psychiatry 64, 318–326 (2007).
Redies, C., Hertel, N. & Hübner, C. A. Cadherins and neuropsychiatric disorders. Brain Res. 1470, 130–144 (2012).
Chang, H. et al. The protocadherin 17 gene affects cognition, personality, amygdala structure and function, synapse development and risk of major mood disorders. Mol. Psychiatry http://dx.doi.org/10.1038/mp.2016.231 (2017).
DeYoung, C. G., Cicchetti, D. & Rogosch, F. A. Moderation of the association between childhood maltreatment and neuroticism by the corticotropin-releasing hormone receptor 1 gene. J. Child Psychol. Psychiatry 52, 898–906 (2011).
Binder, E. B. & Nemeroff, C. B. The CRF system, stress, depression and anxiety—insights from human genetic studies. Mol. Psychiatry 15, 574–588 (2010).
Bulik-Sullivan, B. et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 47, 1236–1241 (2015).
Smith, B. H. et al. Cohort profile: Generation Scotland: Scottish Family Health Study (GS:SFHS). The study, its participants and their potential for genetic research on health and illness. Int. J. Epidemiol. 42, 689–700 (2013).
Burton, R. The Anatomy of Melancholy (eds. Faulkner, T. C., Kiessling, N. K. & Blair, R. L.) (Oxford University Press, Oxford, UK, 1989).
Chang, C. C. et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4, 7 (2015).
Willer, C. J., Li, Y. & Abecasis, G. R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191 (2010).
Boyle, A. P. et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 22, 1790–1797 (2012).
Mi, H. et al. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 45 (D1), D183–D189 (2017).
Gene Ontology Consortium. Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res. 45 (D1), D331–D338 (2017).
Hemani, G. et al. MR-Base: a platform for systematic causal inference across the phenome using billions of genetic associations. Preprint at bioRxiv https://doi.org/10.1101/078972 (2016).
Tobacco and Genetics Consortium. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat. Genet. 42, 441–447 (2010).
Rietveld, C. A. et al. GWAS of 126,559 individuals identifies genetic variants associated with educational attainment. Science 340, 1467–1471 (2013).
Euesden, J., Lewis, C. M. & O’Reilly, P. F. PRSice: polygenic risk score software. Bioinformatics 31, 1466–1468 (2015).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300 (1995).
Acknowledgements
This research has been conducted using the UK Biobank Resource (application numbers 10279 and 4844). Generation Scotland received core support from the Chief Scientist Office of the Scottish Government Health Directorates (CZD/16/6) and the Scottish Funding Council (HR03006). Ethical approval for the GS:SFHS study was obtained from the Tayside Committee on Medical Research Ethics (05/S1401/89 Tayside Committee on Medical Research Ethics A). We are grateful to all the families who took part, the general practitioners and the Scottish School of Primary Care for their help in recruiting them, and the whole Generation Scotland team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists, healthcare assistants and nurses. This work was supported by the University of Edinburgh Centre for Cognitive Ageing and Cognitive Epidemiology, part of the cross-council Lifelong Health and Wellbeing Initiative (MR/K026992/1); funding from the Biotechnology and Biological Sciences Research Council (BBSRC) and the Medical Research Council (MRC) is gratefully acknowledged. This report represents independent research funded in part by the National Institute for Health Research (NIHR) Biomedical Research Centre at South London and the Maudsley NHS Foundation Trust and King’s College London. W.D.H. is supported by a grant from Age UK (Disconnected Mind Project). A.M.M. and I.J.D. are supported by funding from a Wellcome Trust Strategic Award (104036/Z/14/Z).
Author information
Authors and Affiliations
Contributions
M.L. drafted the manuscript with contributions from W.D.H. and I.J.D. G.D., D.C.L., R.E.M., M.J.A. and D.M.H. performed quality control of UK Biobank and/or Generation Scotland data. M.L., G.D., S.P.H. and M.S. analyzed the data. T.-K.C., C.F.-R., W.D.H. and S.E.H. performed or assisted with downstream analysis. C.R.G., C.M.L. and A.M.M. provided critical comments on the manuscript draft and analysis. M.L. and I.J.D. coordinated the work. All authors commented on and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
I.J.D. was a participant in UK Biobank. The other authors declare no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5, Supplementary Tables 1 and 11–13, and Supplementary Note
Supplementary Tables 2–10
Supplementary Tables 2–10
Rights and permissions
About this article
Cite this article
Luciano, M., Hagenaars, S.P., Davies, G. et al. Association analysis in over 329,000 individuals identifies 116 independent variants influencing neuroticism. Nat Genet 50, 6–11 (2018). https://doi.org/10.1038/s41588-017-0013-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-017-0013-8
This article is cited by
-
Polygenic risk for major depression, attention deficit hyperactivity disorder, neuroticism, and schizophrenia are correlated with experience of intimate partner violence
Translational Psychiatry (2024)
-
Effects of sex and gender on the etiologies and presentation of select internalizing psychopathologies
Translational Psychiatry (2024)
-
Cis-meQTL for cocaine use-associated DNA methylation in an HIV-positive cohort show pleiotropic effects on multiple traits
BMC Genomics (2023)
-
Neuroticism mediates the association between childhood abuse and the well-being of community dwelling adult volunteers
BioPsychoSocial Medicine (2023)
-
The contributions of mitochondrial and nuclear mitochondrial genetic variation to neuroticism
Nature Communications (2023)