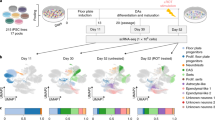
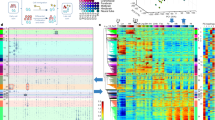
Abstract
Induced pluripotent stem cells (iPSCs), and cells derived from them, have become key tools for modeling biological processes, particularly in cell types that are difficult to obtain from living donors. Here we present a map of regulatory variants in iPSC-derived neurons, based on 123 differentiations of iPSCs to a sensory neuronal fate. Gene expression was more variable across cultures than in primary dorsal root ganglion, particularly for genes related to nervous system development. Using single-cell RNA-sequencing, we found that the number of neuronal versus contaminating cells was influenced by iPSC culture conditions before differentiation. Despite high differentiation-induced variability, our allele-specific method detected thousands of quantitative trait loci (QTLs) that influenced gene expression, chromatin accessibility, and RNA splicing. On the basis of these detected QTLs, we estimate that recall-by-genotype studies that use iPSC-derived cells will require cells from at least 20–80 individuals to detect the effects of regulatory variants with moderately large effect sizes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Change history
03 June 2019
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
References
Itzhaki, I. et al. Modelling the long QT syndrome with induced pluripotent stem cells. Nature 471, 225–229 (2011).
Liu, G.-H. et al. Recapitulation of premature ageing with iPSCs from Hutchinson-Gilford progeria syndrome. Nature 472, 221–225 (2011).
Wainger, B. J. et al. Intrinsic membrane hyperexcitability of amyotrophic lateral sclerosis patient-derived motor neurons. Cell Rep. 7, 1–11 (2014).
Lee, G. et al. Modelling pathogenesis and treatment of familial dysautonomia using patient-specific iPSCs. Nature 461, 402–406 (2009).
Cao, L. et al. Pharmacological reversal of a pain phenotype in iPSC-derived sensory neurons and patients with inherited erythromelalgia. Sci. Transl. Med 8, 335ra56 (2016).
Warren, C. R., Jaquish, C. E. & Cowan, C. A. The NextGen Genetic Association Studies Consortium: a foray into in vitro population genetics. Cell. Stem. Cell. 20, 431–433 (2017).
Young, G. T. et al. Characterizing human stem cell-derived sensory neurons at the single-cell level reveals their ion channel expression and utility in pain research. Mol. Ther. 22, 1530–1543 (2014).
Kilpinen, H. et al. Common genetic variation drives molecular heterogeneity in human iPSCs. Nature 546, 370–375 (2017).
Melé, M. et al. The human transcriptome across tissues and individuals. Science 348, 660–665 (2015).
Soldner, F. et al. Parkinson-associated risk variant in distal enhancer of α-synuclein modulates target gene expression. Nature 533, 95–99 (2016).
Pashos, E. E. et al. Large, diverse population cohorts of hiPSCs and derived hepatocyte-like cells reveal functional genetic variation at blood lipid-associated loci. Cell. Stem. Cell. 20, 558–570 (2017).
Warren, C. R. et al. Induced pluripotent stem cell differentiation enables functional validation of GWAS variants in metabolic disease. Cell. Stem. Cell. 20, 547–557 (2017).
Sala, L., Bellin, M. & Mummery, C. L. Integrating cardiomyocytes from human pluripotent stem cells in safety pharmacology: has the time come? Br. J. Pharmacol. 174, 3749–3765 (2017).
Kiselev, V. Y. et al. SC3: consensus clustering of single-cell RNA-seq data. Nat. Methods. 14, 483–486 (2017).
Newman, A. M. et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 12, 453–457 (2015).
Kléber, M. et al. Neural crest stem cell maintenance by combinatorial Wnt and BMP signaling. J. Cell. Biol. 169, 309–320 (2005).
Ongen, H., Buil, A., Brown, A. A., Dermitzakis, E. T. & Delaneau, O. Fast and efficient QTL mapper for thousands of molecular phenotypes. Bioinformatics. 32, 1479–1485 (2016).
Kumasaka, N., Knights, A. J. & Gaffney, D. J. Fine-mapping cellular QTLs with RASQUAL and ATAC-seq. Nat. Genet. 48, 206–213 (2016).
Li, Y. I. et al. RNA splicing is a primary link between genetic variation and disease. Science 352, 600–604 (2016).
Li, Y. I., Knowles, D. A. & Pritchard, J. K. LeafCutter: annotation-free quantification of RNA splicing. Preprint available at https://www.biorxiv.org/content/early/2016/03/16/044107 (2016).
Buenrostro, J. D., Giresi, P. G., Zaba, L. C., Chang, H. Y. & Greenleaf, W. J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 10, 1213–1218 (2013).
Sheffield, N. C. & Bock, C. LOLA: enrichment analysis for genomic region sets and regulatory elements in R and Bioconductor. Bioinformatics 32, (587–589 (2016).
Lessard, J. et al. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron. 55, 201–215 (2007).
Hunt, S. P., Pini, A. & Evan, G. Induction of c-Fos-like protein in spinal cord neurons following sensory stimulation. Nature 328, 632–634 (1987).
Kohno, T., Moore, K. A., Baba, H. & Woolf, C. J. Peripheral nerve injury alters excitatory synaptic transmission in lamina II of the rat dorsal horn. J. Physiol. (Lond.) 548, 131–138 (2003).
Peters, M. J. et al. Genome-wide association study meta-analysis of chronic widespread pain: evidence for involvement of the 5p15.2 region. Ann. Rheum. Dis. 72, 427–436 (2013).
Pers, T. H., Timshel, P. & Hirschhorn, J. N. SNPsnap: a Web-based tool for identification and annotation of matched SNPs. Bioinformatics. 31, 418–420 (2015).
Gregory, A. P. et al. TNF receptor 1 genetic risk mirrors outcome of anti-TNF therapy in multiple sclerosis. Nature 488, 508–511 (2012).
Probert, L. TNF and its receptors in the CNS: the essential, the desirable and the deleterious effects. Neuroscience. 302, 2–22 (2015).
Spilker, C. & Kreutz, M. R. RapGAPs in brain: multipurpose players in neuronal Rap signalling. Eur. J. Neurosci. 32, 1–9 (2010).
Jordan, J. D. et al. Cannabinoid receptor-induced neurite outgrowth is mediated by Rap1 activation through G(α)o/i-triggered proteasomal degradation of Rap1GAPII. J. Biol. Chem. 280, 11413–11421 (2005).
Melchionda, L. et al. Mutations in APOPT1, encoding a mitochondrial protein, cause cavitating leukoencephalopathy with cytochrome C oxidase deficiency. Am. J. Hum. Genet. 95, 315–325 (2014).
Dianat, N., Steichen, C., Vallier, L., Weber, A. & Dubart-Kupperschmitt, A. Human pluripotent stem cells for modelling human liver diseases and cell therapy. Curr. Gene. Ther. 13, 120–132 (2013).
Smith, B. W. et al. The aryl hydrocarbon receptor directs hematopoietic progenitor cell expansion and differentiation. Blood. 122, 376–385 (2013).
Handel, A. E. et al. Assessing similarity to primary tissue and cortical layer identity in induced pluripotent stem cell-derived cortical neurons through single-cell transcriptomics. Hum. Mol. Genet. 25, 989–1000 (2016).
Hu, B.-Y. et al. Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency. Proc. Natl. Acad. Sci. USA 107, 4335–4340 (2010).
Cacchiarelli, D. et al. Aligning single-cell developmental and reprogramming trajectories identifies molecular determinants of reprogramming outcome. Preprint available at https://www.biorxiv.org/content/early/2017/03/30/122531 (2017).
Musunuru, K. et al. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature 466, 714–719 (2010).
Chambers, S. M. et al. Combined small-molecule inhibition accelerates developmental timing and converts human pluripotent stem cells into nociceptors. Nat. Biotechnol. 30, 715–720 (2012).
Jun, G. et al. Detecting and estimating contamination of human DNA samples in sequencing and array-based genotype data. Am. J. Hum. Genet. 91, 839–848 (2012).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 30, 923–930 (2014).
Hansen, K. D., Irizarry, R. A. & Wu, Z. Removing technical variability in RNA-seq data using conditional quantile normalization. Biostatistics. 13, 204–216 (2012).
Castel, S. E., Levy-Moonshine, A., Mohammadi, P., Banks, E. & Lappalainen, T. Tools and best practices for data processing in allelic expression analysis. Genome. Biol. 16, 195 (2015).
Davis, J. R. et al. An efficient multiple-testing adjustment for eQTL studies that accounts for linkage disequilibrium between variants. Am. J. Hum. Genet. 98, 216–224 (2016).
Sheffield, N. C. et al. Patterns of regulatory activity across diverse human cell types predict tissue identity, transcription factor binding, and long-range interactions. Genome Res. 23, 777–788 (2013).
Danecek, P. et al. The variant call format and VCFtools. Bioinformatics. 27, 2156–2158 (2011).
Acknowledgements
The iPSC lines were generated under the Human Induced Pluripotent Stem Cell Initiative (HIPSCI) funded by a grant from the Wellcome Trust and Medical Research Council (WT098503), supported by the Wellcome Trust (WT098051) and the NIHR/Wellcome Trust Clinical Research Facility. HIPSCI funding was used for sensory neuron RNA-sequencing. We acknowledge Life Science Technologies Corporation as the provider of Cytotune. Pfizer Neuroscience (Pfizer Ltd.) funded neuronal differentiation, functional assays, single-cell RNA-sequencing, and collection and sequencing of dorsal root ganglion samples. The authors gratefully acknowledge N. Kumasaka for help with RASQUAL. We thank F. Merkle for comments on the manuscript. J.S. gratefully acknowledges support from the Wellcome Trust for his PhD studentship. We also thank three anonymous reviewers whose feedback greatly improved this manuscript.
Author information
Authors and Affiliations
Consortia
Contributions
J.S. analyzed data, and J.S. and D.J.G. wrote the manuscript. S.F. performed all differentiations. A. Gutteridge analyzed data. A. Gutteridge, D.J.G., and P.J.W. conceived and supervised the project. H.K. compared eQTLs with GTEx and identified tissue-specific eQTLs. J.R. and M.P. cultured iPSC samples. A.J.K. performed all ATAC-seq. K.A. and A. Goncalves assisted with data analysis. A.W. performed single-cell RNA work and assisted with data analysis. R.F. and C.L.B. performed RNA extraction and quantification. E.I. performed cell culture and Ca2+ flux assays. M.B. assisted with experimental design and Ca2+ flux assays. L.C., S.L., and A.J.L. performed electrophysiology measurements. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
S.F., R.F., C.L.B., A.W., M.B., E.I., L.C., S.L., A.J.L., P.J.W., and A. Gutteridge were all employees of Pfizer at the time the experiments were performed.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Combined Supplementary Figures and Supplementary Note
Supplementary Figures 1–25 and Supplementary Note 1.
Supplementary Tables
Supplementary Tables 1–11.
Supplementary Tables
Supplementary Tables 12–22.
Rights and permissions
About this article
Cite this article
Schwartzentruber, J., Foskolou, S., Kilpinen, H. et al. Molecular and functional variation in iPSC-derived sensory neurons. Nat Genet 50, 54–61 (2018). https://doi.org/10.1038/s41588-017-0005-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-017-0005-8