Abstract
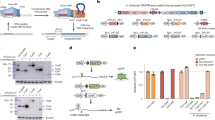
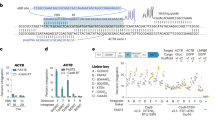
CRISPR-associated transposases (CASTs) enable recombination-independent, multi-kilobase DNA insertions at RNA-programmed genomic locations. However, the utility of type V-K CASTs is hindered by high off-target integration and a transposition mechanism that results in a mixture of desired simple cargo insertions and undesired plasmid cointegrate products. Here we overcome both limitations by engineering new CASTs with improved integration product purity and genome-wide specificity. To do so, we engineered a nicking homing endonuclease fusion to TnsB (named HELIX) to restore the 5′ nicking capability needed for cargo excision on the DNA donor. HELIX enables cut-and-paste DNA insertion with up to 99.4% simple insertion product purity, while retaining robust integration efficiencies on genomic targets. HELIX has substantially higher on-target specificity than canonical CASTs, and we identify several novel factors that further regulate targeted and genome-wide integration. Finally, we extend HELIX to other type V-K orthologs and demonstrate the feasibility of HELIX-mediated integration in human cell contexts.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Sequencing data has been deposited with the NCBI Sequence Read Archive (SRA) under BioProject ID PRJNA889059. All other primary datasets are available in Supplementary Table 4.
References
Hendrie, P. C. & Russell, D. W. Gene targeting with viral vectors. Mol. Ther. 12, 9–17 (2005).
Thomas, C. E., Ehrhardt, A. & Kay, M. A. Progress and problems with the use of viral vectors for gene therapy. Nat. Rev. Genet. 4, 346–358 (2003).
Tellier, M., Bouuaert, C. C. & Chalmers, R. Mariner and the ITm superfamily of transposons. Microbiol. Spectr. 3, 3.2.06 (2015).
van Opijnen, T. & Camilli, A. Transposon insertion sequencing: a new tool for systems-level analysis of microorganisms. Nat. Rev. Microbiol. 11, 435–442 (2013).
Haniford, D. B. & Ellis, M. J. Transposons Tn 10 and Tn 5. Microbiol. Spectr. 3, 3.1.06 (2015).
Plasterk, R. H. A., Izsvák, Z. & Ivics, Z. Resident aliens: the Tc1/ mariner superfamily of transposable elements. Trends Genet. 15, 326–332 (1999).
Wilson, M. H., Coates, C. J. & George, A. L. PiggyBac transposon-mediated gene transfer in human cells. Mol. Ther. 15, 139–145 (2007).
Mali, P. et al. RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013).
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013).
Rouet, P., Smih, F. & Jasin, M. Expression of a site-specific endonuclease stimulates homologous recombination in mammalian cells. Proc. Natl Acad. Sci. USA 91, 6064–6068 (1994).
Wang, H. H. et al. Programming cells by multiplex genome engineering and accelerated evolution. Nature 460, 894–898 (2009).
Wang, H. H. et al. Genome-scale promoter engineering by coselection MAGE. Nat. Methods 9, 591–593 (2012).
Jiang, W., Bikard, D., Cox, D., Zhang, F. & Marraffini, L. A. RNA-guided editing of bacterial genomes using CRISPR–Cas systems. Nat. Biotechnol. 31, 233–239 (2013).
Strecker, J. et al. RNA-guided DNA insertion with CRISPR-associated transposases. Science 365, 48–53 (2019).
Klompe, S. E., Vo, P. L. H., Halpin-Healy, T. S. & Sternberg, S. H. Transposon-encoded CRISPR–Cas systems direct RNA-guided DNA integration. Nature 571, 219–225 (2019).
Vo, P. L. H. et al. CRISPR RNA-guided integrases for high-efficiency, multiplexed bacterial genome engineering. Nat. Biotechnol. 39, 480–489 (2021).
Vo, P. L. H., Acree, C., Smith, M. L. & Sternberg, S. H. Unbiased profiling of CRISPR RNA-guided transposition products by long-read sequencing. Mob. DNA 12, 13 (2021).
Saito, M. et al. Dual modes of CRISPR-associated transposon homing. Cell 184, 2441–2453.e18 (2021).
Strecker, J., Ladha, A., Makarova, K. S., Koonin, E. V. & Zhang, F. Response to comment on “RNA-guided DNA insertion with CRISPR-associated transposases”. Science 368, eabb2920 (2020).
Rubin, B. E. et al. Species- and site-specific genome editing in complex bacterial communities. Nat. Microbiol. 7, 34–47 (2022).
Rybarski, J. R., Hu, K., Hill, A. M., Wilke, C. O. & Finkelstein, I. J. Metagenomic discovery of CRISPR-associated transposons. Proc. Natl Acad. Sci. USA 118, e2112279118 (2021).
May, E. W. & Craig, N. L. Switching from cut-and-paste to replicative Tn7 transposition. Science 272, 401–404 (1996).
Kholodii, G. Y. et al. Four genes, two ends, and a res region are involved in transposition of Tn5053: a paradigm for a novel family of transposons carrying either a mer operon or an integron. Mol. Microbiol. 17, 1189–1200 (1995).
Hickman, A. B. et al. Unexpected structural diversity in DNA recombination. Mol. Cell 5, 1025–1034 (2000).
Xu, S. Sequence-specific DNA nicking endonucleases. Biomol. Concepts 6, 253–267 (2015).
Gasiunas, G., Barrangou, R., Horvath, P. & Siksnys, V. Cas9–crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl Acad. Sci. USA 109, E2579–E2586 (2012).
Jinek, M. et al. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012).
Xu, S. & Gupta, Y. K. Natural zinc ribbon HNH endonucleases and engineered zinc finger nicking endonuclease. Nucleic Acids Res. 41, 378–390 (2013).
McConnell Smith, A. et al. Generation of a nicking enzyme that stimulates site-specific gene conversion from the I-AniI LAGLIDADG homing endonuclease. Proc. Natl Acad. Sci. USA 106, 5099–5104 (2009).
Niu, Y., Tenney, K., Li, H. & Gimble, F. S. Engineering variants of the I-SceI homing endonuclease with strand-specific and site-specific DNA-nicking activity. J. Mol. Biol. 382, 188–202 (2008).
Kong, S., Liu, X., Fu, L., Yu, X. & An, C. I-PfoP3I: a novel nicking HNH homing endonuclease encoded in the group I intron of the DNA polymerase gene in Phormidium foveolarum phage Pf-WMP3. PLoS ONE 7, e43738 (2012).
Landthaler, M. & Shub, D. A. The nicking homing endonuclease I-BasI is encoded by a group I intron in the DNA polymerase gene of the Bacillus thuringiensis phage Bastille. Nucleic Acids Res. 31, 3071–3077 (2003).
Shen, Y. et al. Structural basis for DNA targeting by the Tn7 transposon. Nat. Struct. Mol. Biol. 29, 143–151 (2022).
Stoddard, B. L. Homing endonucleases from mobile group I introns: discovery to genome engineering. Mob. DNA 5, 7 (2014).
Takeuchi, R., Certo, M., Caprara, M. G., Scharenberg, A. M. & Stoddard, B. L. Optimization of in vivo activity of a bifunctional homing endonuclease and maturase reverses evolutionary degradation. Nucleic Acids Res. 37, 877–890 (2009).
Querques, I., Schmitz, M., Oberli, S., Chanez, C. & Jinek, M. Target site selection and remodelling by type V CRISPR–transposon systems. Nature 599, 497–502 (2021).
Park, J.-U., Tsai, A. W.-L., Chen, T. H., Peters, J. E. & Kellogg, E. H. Mechanistic details of CRISPR-associated transposon recruitment and integration revealed by cryo-EM. Proc. Natl. Acad. Sci. USA 119, e2202590119 (2022).
Tenjo-Castaño, F. et al. Structure of the TnsB transposase-DNA complex of type V-K CRISPR-associated transposon. Nat. Commun. 13, 5792 (2022).
Liu, R., Qiu, J., Finger, L. D., Zheng, L. & Shen, B. The DNA-protein interaction modes of FEN-1 with gap substrates and their implication in preventing duplication mutations. Nucleic Acids Res. 34, 1772–1784 (2006).
Scalley-Kim, M., McConnell-Smith, A. & Stoddard, B. L. Coevolution of a homing endonuclease and its host target sequence. J. Mol. Biol. 372, 1305–1319 (2007).
Gilpatrick, T. et al. Targeted nanopore sequencing with Cas9-guided adapter ligation. Nat. Biotechnol. 38, 433–438 (2020).
Park, J.-U. et al. Structural basis for target site selection in RNA-guided DNA transposition systems. Science 373, 768–774 (2021).
Schmitz, M., Querques, I., Oberli, S., Chanez, C. & Jinek, M. Structural basis for RNA-mediated assembly of type V CRISPR-associated transposons. Preprint at BioRxiv https://doi.org/10.1101/2022.06.17.496590 (2022).
Mizuno, N. et al. MuB is an AAA+ ATPase that forms helical filaments to control target selection for DNA transposition. Proc. Natl Acad. Sci. USA 110, E2441–E2450 (2013).
Skelding, Z., Queen-Baker, J. & Craig, N. L. Alternative interactions between the Tn7 transposase and the Tn7 target DNA binding protein regulate target immunity and transposition. EMBO J. 22, 5904–5917 (2003).
Stellwagen, A. E. & Craig, N. L. Avoiding self: two Tn7-encoded proteins mediate target immunity in Tn7 transposition. EMBO J. 16, 6823–6834 (1997).
Kolter, R., Inuzuka, M. & Helinski, D. R. Trans-complementation-dependent replication of a low molecular weight origin fragment from plasmid R6K. Cell 15, 1199–1208 (1978).
Metcalf, W. W., Jiang, W. & Wanner, B. L. Use of the rep technique for allele replacement to construct new Escherichia coli hosts for maintenance of R6K gamma origin plasmids at different copy numbers. Gene 138, 1–7 (1994).
Klompe, S. E. et al. Evolutionary and mechanistic diversity of Type I-F CRISPR-associated transposons. Mol. Cell 82, 616–628.e5 (2022).
Jonathan Strecker, Feng Zhang, Alim Ladha. CRISPR-associated transposase systems and methods of use thereof. International patent WO2020131862A1 (2020).
Wu, Z. & Chaconas, G. Flanking host sequences can exert an inhibitory effect on the cleavage step of the in vitro mu DNA strand transfer reaction. J. Biol. Chem. 267, 9552–9558 (1992).
Krüger, R. & Filutowicz, M. Dimers of pi protein bind the A+T-rich region of the R6K gamma origin near the leading-strand synthesis start sites: regulatory implications. J. Bacteriol. 182, 2461–2467 (2000).
Harshey R. M, Transposable phage Mu. Microbiol. Spectr. 2, 2.5.31 (2014).
Chalmers, R., Guhathakurta, A., Benjamin, H. & Kleckner, N. IHF modulation of Tn10 transposition: sensory transduction of supercoiling status via a proposed protein/DNA molecular spring. Cell 93, 897–908 (1998).
Swingle, B., O’Carroll, M., Haniford, D. & Derbyshire, K. M. The effect of host-encoded nucleoid proteins on transposition: H-NS influences targeting of both IS903 and Tn10. Mol. Microbiol. 52, 1055–1067 (2004).
Zayed, H., Izsvák, Z., Khare, D., Heinemann, U. & Ivics, Z. The DNA-bending protein HMGB1 is a cellular cofactor of Sleeping Beauty transposition. Nucleic Acids Res. 31, 2313–2322 (2003).
Filutowicz, M. & Appelt, K. The integration host factor of Escherichia coli binds to multiple sites at plasmid R6K gamma origin and is essential for replication. Nucleic Acids Res. 16, 3829–3843 (1988).
Sharpe, P. L. & Craig, N. L. Host proteins can stimulate Tn7 transposition: a novel role for the ribosomal protein L29 and the acyl carrier protein. EMBO J. 17, 5822–5831 (1998).
Parks, A. R. et al. Transposition into replicating DNA occurs through interaction with the processivity factor. Cell 138, 685–695 (2009).
Strecker, J. et al. Engineering of CRISPR–Cas12b for human genome editing. Nat. Commun. 10, 212 (2019).
Xu, X. et al. Engineered miniature CRISPR–Cas system for mammalian genome regulation and editing. Mol. Cell 81, 4333–4345 (2021).
Kim, D. Y. et al. Efficient CRISPR editing with a hypercompact Cas12f1 and engineered guide RNAs delivered by adeno-associated virus. Nat. Biotechnol. 40, 94–102 (2022).
Anzalone, A. V. et al. Programmable deletion, replacement, integration and inversion of large DNA sequences with twin prime editing. Nat. Biotechnol. 40, 731–740 (2022).
Ioannidi, E. I. et al. Drag-and-drop genome insertion without DNA cleavage with CRISPR-directed integrases. Preprint at BioRxiv https://doi.org/10.1101/2021.11.01.466786 (2021).
Li, H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100 (2018).
Rohland, N. & Reich, D. Cost-effective, high-throughput DNA sequencing libraries for multiplexed target capture. Genome Res 22, 939–946 (2012).
Kleinstiver, B. P. et al. Engineered CRISPR–Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing. Nat. Biotechnol. 37, 276–282 (2019).
Acknowledgements
We thank L. T. Hille, C. R. R. Alves, and R. A. Silverstein for suggestions about the manuscript, P. M. Boone for assistance with ddPCR, and B. L. Stoddard for advice. C.J.T. was supported by a National Science Foundation Graduate Research Fellowship grant number 2020295403. B.P.K. was supported by a Mass General Hospital Howard M. Goodman Fellowship.
Author information
Authors and Affiliations
Contributions
C.J.T conceived of the idea, designed and performed experiments, and wrote the manuscript draft. B.O. conducted experiments for ShoHELIX and cargo size comparisons. B.P.K supervised the study and contributed to experimental design. C.J.T and B.P.K wrote and revised the manuscript with input from B.O.
Corresponding author
Ethics declarations
Competing interests
C.J.T. and B.P.K are inventors on patents and/or patent applications filed by Massachusetts General Brigham that describe genome-engineering technologies. B.P.K. is a consultant for EcoR1 capital, and is an advisor to Acrigen Biosciences, Life Edit Therapeutics, and Prime Medicine. B.O. declares no competing interests.
Peer review
Peer review information
Nature Biotechnology thanks Sheng Yang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Table of Contents for Supplementary Information, Titles of Supplementary Tables 1–4, Supplementary Notes 1–6, Supplementary Figs 1–13, and Supplementary References.
Supplementary Table
Supplementary Tables 1–4 separated by tabs.
Supplementary Data
PDF file of uncropped gel images.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Tou, C.J., Orr, B. & Kleinstiver, B.P. Precise cut-and-paste DNA insertion using engineered type V-K CRISPR-associated transposases. Nat Biotechnol 41, 968–979 (2023). https://doi.org/10.1038/s41587-022-01574-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41587-022-01574-x
This article is cited by
-
Precise genome-editing in human diseases: mechanisms, strategies and applications
Signal Transduction and Targeted Therapy (2024)
-
Targeted DNA integration in human cells without double-strand breaks using CRISPR-associated transposases
Nature Biotechnology (2024)
-
Enhancing prime editor activity by directed protein evolution in yeast
Nature Communications (2024)
-
Bacterial genome engineering using CRISPR-associated transposases
Nature Protocols (2024)
-
CRISPR technologies for genome, epigenome and transcriptome editing
Nature Reviews Molecular Cell Biology (2024)