Abstract
Spatial transcriptomics and proteomics provide complementary information that independently transformed our understanding of complex biological processes. However, experimental integration of these modalities is limited. To overcome this, we developed Spatial PrOtein and Transcriptome Sequencing (SPOTS) for high-throughput simultaneous spatial transcriptomics and protein profiling. Compared with unimodal measurements, SPOTS substantially improves signal resolution and cell clustering and enhances the discovery power in differential gene expression analysis across tissue regions.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
All raw data generated in the present study have been deposited to the GEO with accession no. GSE198353. High-resolution images of the tissues used in the present study are available at the Figshare website (https://figshare.com/account/home#/projects/143019). Source data are provided with this paper.
Code availability
To enable easy access to the SPOTS computational pipeline, the auxiliary R package ‘spots’ can be found on the CRAN (https://CRAN.R-project.org/package=spots) and GitHub (https://github.com/stevexniu/spots) websites. The R markdown files for SPOTS data analysis are included in Supplementary Notes 3 and 4 and also on Github (https://github.com/stevexniu/SPOTS-paper).
References
Tabula Muris Consortium et al. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 562, 367–372 (2018).
Azizi, E. et al. Single-cell map of diverse immune phenotypes in the breast tumor microenvironment. Cell 174, 1293–1308.e36 (2018).
Zhao, T. et al. Spatial genomics enables multi-modal study of clonal heterogeneity in tissues. Nature 601, 85–91 (2022).
Chen, A. et al. Spatiotemporal transcriptomic atlas of mouse organogenesis using DNA nanoball-patterned arrays. Cell 185, 1777–1792.e21 (2022).
Fu, X. et al. Continuous polony gels for tissue mapping with high resolution and RNA capture efficiency. Preprint at bioRxiv https://doi.org/10.1101/2021.03.17.435795 (2021).
Vickovic, S. et al. High-definition spatial transcriptomics for in situ tissue profiling. Nat. Methods 16, 987–990 (2019).
Stickels, R. R. et al. Highly sensitive spatial transcriptomics at near-cellular resolution with Slide-seqV2. Nat. Biotechnol. 39, 313–319 (2021).
Gut, G., Herrmann, M. D. & Pelkmans, L. Multiplexed protein maps link subcellular organization to cellular states. Science 361, eaar7042 (2018).
Radtke, A. J. et al. IBEX: a versatile multiplex optical imaging approach for deep phenotyping and spatial analysis of cells in complex tissues. Proc. Natl Acad. Sci. USA 117, 33455–33465 (2020).
Angelo, M. et al. Multiplexed ion beam imaging of human breast tumors. Nat. Med. 20, 436–442 (2014).
Giesen, C. et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat. Methods 11, 417–422 (2014).
Walch, A., Rauser, S., Deininger, S.-O. & Höfler, H. MALDI imaging mass spectrometry for direct tissue analysis: a new frontier for molecular histology. Histochem. Cell Biol. 130, 421–434 (2008).
Goltsev, Y. et al. Deep profiling of mouse splenic architecture with CODEX multiplexed imaging. Cell 174, 968–981.e15 (2018).
Govek, K. W. et al. Single-cell transcriptomic analysis of mIHC images via antigen mapping. Sci. Adv. 7, eabc5464 (2021).
Jiang, S. et al. Combined protein and nucleic acid imaging reveals virus-dependent B cell and macrophage immunosuppression of tissue microenvironments. Immunity https://doi.org/10.1016/j.immuni.2022.03.020 (2022).
Liu, Y. et al. High-spatial-resolution multi-omics sequencing via deterministic barcoding in tissue. Cell 183, 1665–1681.e18 (2020).
Guilliams, M. et al. Spatial proteogenomics reveals distinct and evolutionarily conserved hepatic macrophage niches. Cell 185, 379–396.e38 (2022).
Vickovic, S. et al. SM-Omics is an automated platform for high-throughput spatial multi-omics. Nat. Commun. 13, 795 (2022).
Ståhl, P. L. et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 353, 78–82 (2016).
Stoeckius, M. et al. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 14, 865–868 (2017).
Lim, H. K. & O’Neill, H. C. Identification of stromal cells in spleen which support myelopoiesis. Front. Cell Dev. Biol. 7, 1 (2019).
Cheng, H.-W. et al. Origin and differentiation trajectories of fibroblastic reticular cells in the splenic white pulp. Nat. Commun. 10, 1739 (2019).
Béguelin, W. et al. EZH2 is required for germinal center formation and somatic EZH2 mutations promote lymphoid transformation. Cancer Cell 23, 677–692 (2013).
Liu, Y., Beyer, A. & Aebersold, R. On the dependency of cellular protein levels on mRNA abundance. Cell 165, 535–550 (2016).
Mimitou, E. P. et al. Scalable, multimodal profiling of chromatin accessibility, gene expression and protein levels in single cells. Nat. Biotechnol. 39, 1246–1258 (2021).
Chávez-Galán, L., Olleros, M. L., Vesin, D. & Garcia, I. Much more than M1 and M2 macrophages, there are also CD169+ and TCR+ macrophages. Front. Immunol. 6, 263 (2015).
Ravishankar, B. et al. Marginal zone CD169+ macrophages coordinate apoptotic cell-driven cellular recruitment and tolerance. Proc. Natl Acad. Sci. USA 111, 4215–4220 (2014).
Nagelkerke, S. Q. et al. Red pulp macrophages in the human spleen are a distinct cell population with a unique expression of Fc-γ receptors. Blood Adv. 2, 941–953 (2018).
Biancalani, T. et al. Deep learning and alignment of spatially resolved single-cell transcriptomes with Tangram. Nat. Methods 18, 1352–1362 (2021).
Valdés-Mora, F. et al. Single-cell transcriptomics reveals involution mimicry during the specification of the basal breast cancer subtype. Cell Rep. 35, 108945 (2021).
Keren, L. et al. A structured tumor-immune microenvironment in triple negative breast cancer revealed by multiplexed ion beam imaging. Cell 174, 1373–1387.e19 (2018).
Franklin, R. A. et al. The cellular and molecular origin of tumor-associated macrophages. Science 344, 921–925 (2014).
Elosua-Bayes, M., Nieto, P., Mereu, E., Gut, I. & Heyn, H. SPOTlight: seeded NMF regression to deconvolute spatial transcriptomics spots with single-cell transcriptomes. Nucleic Acids Res. 49, e50 (2021).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29 (2021).
Nam, A. S. et al. Somatic mutations and cell identity linked by genotyping of transcriptomes. Nature 571, 355–360 (2019).
Hudson, W. H. & Sudmeier, L. J. Localization of T cell clonotypes using spatial transcriptomics. Preprint at bioRxiv https://doi.org/10.1101/2021.08.03.454999 (2021).
Dhainaut, M. et al. Spatial CRISPR genomics identifies regulators of the tumor microenvironment. Cell 185, 1223–1239.e20 (2022).
Roelli, P., Flynn, B. & Gui, G. Hoohm/CITE-seq-Count: 1.4.2. Zenodo https://zenodo.org/record/2590196#.Y07yOy8Ro8U (2019).
Ord, J. K. & Getis, A. Local spatial autocorrelation statistics: distributional issues and an application. Geogr. Anal. 27, 286–306 (2010).
Anselin, L. Local indicators of spatial association—LISA. Geogr. Anal. 27, 93–115 (2010).
Getis, A. & Ord, J. K. The analysis of spatial association by use of distance statistics. Geogr. Anal. 24, 189–206 (2010).
Middleton, L. & Sivaswamy, J. Edge detection in a hexagonal-image processing framework. Image Vis. Comput. 19, 1071–1081 (2001).
Haghverdi, L., Lun, A. T. L., Morgan, M. D. & Marioni, J. C. Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat. Biotechnol. 36, 421–427 (2018).
Moran, P. A. P. Notes on continuous stochastic phenomena. Biometrika 37, 17–23 (1950).
Wartenberg, D. Multivariate spatial correlation: A method for exploratory geographical analysis. Geogr. Anal. 17, 263–283 (2010).
Czaplewski, R. L. Expected value and variance of Moran’s bivariate spatial autocorrelation statistic for a permutation test. Rocky Mountain Forest and Range Experiment Station (US Department of Agriculture, Forest Service, 1993).
Lee, S.-I. Developing a bivariate spatial association measure: an integration of Pearson’s r and Moran’s I. J. Geogr. Syst. 3, 369–385 (2001).
Frost, H. R. Eigenvectors from eigenvalues sparse principal component analysis. J. Comput. Graph. Stat. 31, 486–501 (2022).
Niu, X. spots: Spatial Component Analysis. Zenodo https://doi.org/10.5281/zenodo.6918175 (2022).
Korotkevich, G. et al. Fast gene set enrichment analysis. Preprint at bioRxiv https://doi.org/10.1101/060012 (2016).
Acknowledgements
We thank J. Chew and Y. Yin from 10x Genomics Headquarters for critical discussions and K. Ganapathy for helping coordinate 10x Genomics and Weill Cornell interactions. X.N. can list himself first in authors in his CV. D.A.L. is supported by the Burroughs Wellcome Fund Career Award for Medical Scientists (no. 1014689-01), Valle Scholar Award (no. VS-2020-31), Leukemia Lymphoma Scholar Award (no. 1373-21), the Sontag Foundation Distinguished Scientist Award (no. SFI 203261-02), the Mark Foundation Emerging Leader Award (no. 21-042-ELA) and the National Institutes of Health Director’s New Innovator Award (no. DP2-CA239065). This work was supported by the CEGS award (no. RM1 HG011014) and Emerson (NPT Charitable grant no. 584001).
Author information
Authors and Affiliations
Contributions
N.B.-C., X.N., M.S. and D.A.L. conceived the project and wrote the manuscript with input from all authors. N.B.-C., M.S., A.D.S., J.S., M.S.J., C.M.S., P.M. and P.R. performed the experiments. X.N. performed the bioinformatic analyses. C.P. edited the manuscript. D.A.L. supervised the study. All authors reviewed and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
M.S., P.M. and P.R. are current employees of 10x Genomics, Sweden. D.A.L. has served as a consultant for Abbvie, AstraZeneca and Illumina, and is on the Scientific Advisory Board or equity holder of Mission Bio, Pangea, Alethiomics and C2i Genomics. D.A.L. has also received previous research funding from BMS, 10x Genomics, Ultima Genomics and Illumina unrelated to the current manuscript. The other authors declare no competing interests.
Peer review
Peer review information
Nature Biotechnology thanks Shalev Itzkovitz, Andreas Moor and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Workflow schematic of SPOTS.
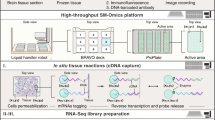
(Step 1) Tissue sections are mounted onto Visium slide, fixed for 10 min, and immunostained (Step 2) with fluorescent and TotalSeq-A antibodies (same clones) for 90 min in the presence of poly-T blocking oligos that prevent non-specific ADTs binding to slide surface. After washes, tissue is scanned under a scanning microscope to capture the tissue structure and fiducial spots (Step 3). The tissue is then permeabilized (Step 4), and RNA diffuses onto the poly-T oligos on the spatial barcodes. Following tissue digestion (Step 5), and template switched RT-PCR (Step 6), gene expression and ADT libraries are generated to preserve associated spatial barcodes and RNA sequence/ADT antibody barcodes (Step 7). Gene expression and ADT libraries are sequenced, and data are integrated using our designated pipeline (Step 8).
Extended Data Fig. 2 SPOTS library preparation and optimizations.
(a) Illustration of the structure of a barcoded oligo, including Read 1, spatial barcode (SB), unique molecular identifier (UMI), and poly-T (T30) sequences, attached to the Visium slide. (b) Illustration of the structure of an ADT-conjugated antibody, including a PCR handle (Handle), an antibody barcode (AB), and poly-A (A30). (c) Schematic of SPOTS following second strand synthesis. ADT and mRNA location is recorded through spatial barcodes on the Visium slide. (d) After cDNA amplification, ADT and cDNA libraries can be separated and prepared for sequencing to produce indexed ADT libraries (top panel, BioA) and gene expression cDNA libraries (bottom panel, BioA). (e) IF analysis using CD29 antibodies (TotalSeq-A clone) in spleen tissues that were fixed for 10 min at 25 °C using 100% Methanol (MeOH, top) or 1% PFA (bottom). Scale bar 200 μm. (f) Normalized ADT levels of indicated antibodies (left) and cDNA libraries (right, BioA) using tissue permeabilization vs. tissue removal enzyme and SDS. (g) Poly-dT blocking oligo length and wash temperature titration. Titration of different poly-dT blocking oligo lengths and their ability to hinder binding of dual-tagged antibodies (ADT + fluorophore) to poly-A surface probes at either 4 °C or 37 °C. Note the binding differences at the different temperatures for dT20. (h) IF using dual-tagged antibodies (ADT + fluorophore) of CD29 (green) and CD4 (red) in mouse spleens in the presence or absence of 20 µM poly-dT20 blocking oligos. Note for reduction of non-specific bindings in tissue-free areas. Scale bar 600 μm.
Extended Data Fig. 3 Normalized ADT levels in spleens.
(a) Normalized ADT levels of all 21 lineage and functional markers surface proteins in two spleen samples. Markers include; B cells (CD19, CD20, CD220, IgD, IgM, CD38), T cells (CD3, CD8, CD4), scavenging and antigen-presenting macrophages (CD169, F4/80, CD163, CD68, CD11b), granulocytes (Ly6G, Ly6C), endothelial cells (CD31, MadCAM1), and fibroblasts (CD29, CD105). (b) Immunofluorescence staining for CD29 (green) and CD4 (red) in spleen tissues from a, using fluorophore-ADT dual-tagged antibodies. Scale bar 300 μm. (c) Serial histological sections of mouse spleen (unstimulated) stained for H&E, B cells (B220), and T cells (CD8), demonstrating the typical spatial cell organization. Scale bar 300 μm.
Extended Data Fig. 4 Reproducibility and quality controls of SPOTS spleen data.
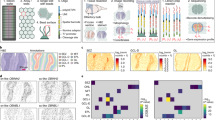
(a) Correlation of normalized ADT levels and mRNA expressions between two biological replicates of mouse spleen. The blue lines represent linear regression lines and 95% confident intervals are shown as gray shades. (b) Correlation between mRNA and different ADT levels (UMI) at single spatial barcode level for B cell, T cell, and macrophage enriched clusters across spatial barcodes in two biological replicates. The blue lines represent linear regression lines and 95% confident intervals are shown as gray shades. (c) Boxplot showing mRNA expression levels of germinal center (GC) specific genes (GSE12366; n = 163 genes) as a function of the distance from the center of GCs. The boxes were colored by the physical distance from the center of GCs as shown in Fig. 1f. Each boxplot ranges from the first and third quartiles with median shown as middle line, and the whiskers represents 1.5 times the interquartile range. (d) Cell-type deconvolution based on ADTs (Z-score) of each spatial barcode overlaid onto the spleen tissue. Magnified areas are shown in Fig. 1h.
Extended Data Fig. 5 Comparative performance analyses between SPOTS, Visium- alone, and Guilliams et al.
(a) Total detected genes and UMIs across spatial barcodes in biological replicates of spleens using SPOTS or Visium alone (n = 2 each). Sequencing depth: SPOTS rep A ~175 K reads per spot, SPOTS rep B ~155 K reads per spot, Visium rep A ~124 K reads per spot, Visium rep B ~110 K reads per spot. Each boxplot ranges from the first and third quartiles with median shown as middle line, and the whiskers represents 1.5 times the interquartile range. (b) Sequencing saturation curves for RNA of the two replicates of mouse spleen using SPOTS or Visium alone. (c) Correlation between mRNA and ADT UMI counts (left panel) in spleens using SPOTS. Correlation of total mRNA UMI counts in spatial barcodes using SPOTS (Y-axis) and Visium only (X-axis). The blue lines represent linear regression lines and 95% confident intervals are shown as gray shades. (d) Correlation of gene expression signatures in the indicated cell clusters as detected by SPOTS (Y-axis) and Visium alone (X-axis). (e) Total ADT UMI counts across the tissue spatial barcodes using 93 or 21 TotalSeq-A antibodies (Guilliams in mouse liver or SPOTS in mouse spleen, respectively). Each boxplot ranges from the first and third quartiles with median shown as middle line, and the whiskers represents 1.5 times the interquartile range. (f) Correlation map between several ADT markers of the same cell type in mouse liver (Guilliams et al.) and mouse spleen (SPOTS). ADTs: T-cells (CD8, CD4, CD3), B-cells (CD19, B220, IgD), and macrophages (F4/80, CD68, CD163). (g) Visualization of normalized ADTs of macrophages from panel b in liver (Guilliams et al.) and spleen (SPOTS).
Extended Data Fig. 6 Performance comparison of SPOTS to SM-Omics.
(a) Total mRNA and ADT counts across spatial barcodes recovered in SM-Omics and SPOTS in biological replicates of spleens. (b) Total ADT counts of F4/80 (macrophages) and IgD (B cells) across biological replicates of spleens. (c) Correlations between IgD and F4/80 (normalized ADT counts) for each spatial barcode in spleen by SPOTS and SM-Omics. Note the anti-correlation between F4/80+ macrophage-enriched (Red pulp) and IgD+ B cell-enriched (B follicle) regions in the SPOTS analyses. (d) Differentially expressed ADT levels (Z-score) for each cluster of spatial barcodes in spleen. Left panel: SPOTS, right panel: SM-Omics (e) Heatmap showing differentially expressed mRNAs (Z-score) in each cluster of spatial barcodes in splenic tissues. Left panel: SPOTS, right panel: SM-Omics (f) Visualization of normalized ADT expression of macrophages (F4/80) and B-cells (IgD) in spleens using SPOTS (left) and SM-Omics (right).
Extended Data Fig. 7 Breast cancer tissue architecture and optimizations.
(a) Immunohistochemistry (IHC) analysis of CD8 (T cells) and IBA-1 (macrophages) in mammary tumor section from MMTV-PyMT model demonstrating the typical abundance of T cells and tumor-associated macrophages in breast tumors. Right, schematic representation of Visium spatial capture spot on tissue and cell type abundances within. (b) Fluorescence imaging of TRITC labeled cDNA from mouse breast tumors following 5 or 10 minutes with 5X or 10X saponin or 0.1% Triton pre-staining permeabilization conditions. Dotted lines outline tissue borders.
Extended Data Fig. 8 Gene expression analysis of housekeeping genes in SPOTS and Visium cDNA libraries and comparative performance analysis between SPOTS and Visium-alone in breast cancer tissue.
(a) Images of serial sections of 3 biological replicates of mammary tumors using Visium protocol (H&E staining images) or SPOTS protocol (IF images of EpCAM). (b) Gene expression cycle threshold (CT) values of the indicated housekeeping genes using cDNA generated by Visium or SPOTS protocols from panel a. Each boxplot ranges from the first and third quartiles with median shown as middle line, and the whiskers represents 1.5 times the interquartile range. (c) Total detected genes across spatial barcodes in biological replicates of mammary tumors using SPOTS (n = 3) or Visium alone (n = 4). Sequencing depths for all Visium and SPOTS samples ~50 K reads per spot. Each boxplot ranges from the first and third quartiles with median shown as middle line, and the whiskers represents 1.5 times the interquartile range. (d) Sequencing saturation curves for RNA of the biological replicates from panel c.
Extended Data Fig. 9 Spatial mRNA and ADT levels in breast cancer TME.
(a) Normalized ADT levels (Z-score) of selected markers in a tissue section of MMTV-PyMT breast cancer. (b) Correlation between normalized mRNA expression and ADT levels of the indicated surface markers (EpCAM, PDPN, SCA-1, MHC-II). Pearson’s correlation coefficients were calculated on cluster-level and are indicated in the boxed labels. The blue lines represent linear regression lines, and 95% confident intervals are shown as gray shades. (c) Average normalized mRNA expressions of key cell-type marker genes (GSE158677) in each spatial cluster. (d) H&E staining and IHC analysis of KRT18 (tumor cells), CD8 (T cells) and IBA1 (macrophages) in serial sections of the MMTV-PyMT breast cancer model, demonstrating the typical abundances of tumor cells, T cells, and macrophages in mammary tumors. Scale bar 500 μm. (e) Normalized mRNA and ADT expression levels of Epcam and Ptprc along with their corresponding IF signals in breast cancer tissue. (f) Pearson’s correlation analysis between total UMI mRNA or UMI ADT counts and IF intensities of the indicated genes across the spatial barcodes. Pearson’s correlation coefficients are shown; mRNA/IF (top) and ADT/IF (bottom). The blue lines represent linear regression lines, and 95% confident intervals are shown as gray shades. P-values were calculated with two-sided Pearson’s correlation tests and exact p-values were labeled on each panel.
Extended Data Fig. 10 IF analysis using TotalSeq-A clones in breast cancer.
(a) IF analysis of EpCAM and DAPI to visualize tissue architecture along with F4/80 and MHC-II to highlight tumor-associated macrophages (TAMs). Scale bar 500 μm. (b) IF analysis of EpCAM and DAPI to visualize tissue architecture along with CCR2 and MHC-II to highlight two populations of TAMs (MHC-II + CCR2-/MHC-II + CCR2 + ). Scale bar 500 μm. (c) IF analysis of EpCAM and DAPI to visualize tissue architecture along with CD4, CD8 to mark the two populations of tumor-infiltrating T cells. Scale bar 500 μm. (d) IF analysis of EpCAM and DAPI to visualize tissue architecture and CD31 to visualize blood vessels. Scale bar 500 μm.
Supplementary information
Supplementary Information
Supplementary Notes 1–4.
Supplementary Table
Supplementary Tables 1–5.
Source data
Source Data Extended Data Fig. 8
Quantitastive PCR CT values of housekeeping genes in Visium_vs_SPOTS (breast cancer).
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ben-Chetrit, N., Niu, X., Swett, A.D. et al. Integration of whole transcriptome spatial profiling with protein markers. Nat Biotechnol 41, 788–793 (2023). https://doi.org/10.1038/s41587-022-01536-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41587-022-01536-3
This article is cited by
-
The interaction between ageing and Alzheimer's disease: insights from the hallmarks of ageing
Translational Neurodegeneration (2024)
-
Spatial multi-omics: novel tools to study the complexity of cardiovascular diseases
Genome Medicine (2024)
-
Decoder-seq enhances mRNA capture efficiency in spatial RNA sequencing
Nature Biotechnology (2024)
-
Multiplex protein imaging in tumour biology
Nature Reviews Cancer (2024)
-
CellCharter reveals spatial cell niches associated with tissue remodeling and cell plasticity
Nature Genetics (2024)